-
Hepatitis B virus (HBV) infection is a global health problem. HBV has been classified into eight genotypes (A-H) based on genome sequence divergence, and moreover, the eight genotypes can be further subdivided into subgenotypes with distinct ethnic or geographical origin. In recent years, there is increasing evidence which indicates the correlation between HBV genotypes (or subgenotypes) and the transmission modes of the virus as well as its prognosis and prevention, the choice of antiviral therapies. Therefore, the analysis of the distribution of HBV genotypes (or subgenotypes) among different regions or ethnic groups and the definition on the epidemic tendency of different genotypes (or subgenotypes) at the molecular level are of great significance for the prevention and treatment of hepatitis B.
Dali, in Yunnan Province, is a district inhabited by ethnic minorities in which Bai nationality accounts for the main proportion of the population. To date, a study on HBV genotypes and subgenotypes of this group has not been reported. Therefore, we carried out a preliminary investigation on the HBV genotypes and subgenotypes among the population of Bai nationals in this district with an aim of determining their distribution within this group.
HTML
-
A total 100 serum samples were obtained from chronically HBV-infected patients of Bai nationality who visited the Dali Bai Autonomous District Hospital or Dali City Hospital or Dali maternal & child health institute. Serum HBsAg in all patients was positive for at least 6 months. In addition, all patients were not HCV infected. The study included 65 males and 35 females aged from 0.75~78 years (average: 29.3 ± 14.3 years). The standard HBV markers, including HBsAg, HBsAb, HBeAg, HBe Ab and HBcAb, were tested with commercial kits (ELISA; Kehua Bio-engineering Co. Ltd, Shanghai, China).
-
All the primers in this study were synthesized by Sangon (Shanghai). The restriction endonucleases (BstEⅡ, Bcn Ⅰ, Hpa Ⅰ, Stu Ⅰ) for RFLP were purchased from MBI Fermentas.
-
HBV DNA was extracted from serum that had been stored at -80℃ using a rapid and simple method described previously (9).
-
The primers used in this study were designed according to Naito's study (5). The sequences of PCR primers are shown in Table 1. The method can distinguish genotype A through F. HBV genotypes were identified by nested PCR. The first-round PCR was carried in a tube containing 40μL of a reaction buffer made up of the following components: 6μL of Template, 200 μmol/L of dNTPs, 0.2 μmol/L of each universal primer (P1 and S1), 1.5 U of Taq polymerase. The cycle parameters were an initial 10 min denaturation at 94℃, followed by 40 cycles consisting of 94℃ for 20 s, 55℃ for 20 s, and 72℃ for 60 s. Strand synthesis was completed at 72℃ extension for 7 min. The second-round PCR was divided into two groups (A and B). 2μL of the first PCR products for template was used for the second-round PCR under the same reaction system but with the primers of group A (B2、BA1R、BB1R、BC1R) and group B (B2R、BD1、BE1、BF1). Both group A and B were performed within the same cycle parameters: preheating at 95℃ for 10 min, 20 cycles of amplification at 94℃ for 20 s, 58℃ for 20 s, and 72℃ for 30 s, then an additional 20 cycles of 94℃ for 20 s, 60℃ for 20 s, and 72℃ for 30 s, and finally, extension at 72℃ for 7 min. The two different second-round PCR products from one sample were separately electrophoresed on a 2.5% agarose gel, stained with ethidium bromide and visualized under UV light.

Table 1. Primer sequence of HBV genotyping
HBV genotype was determined by the size of amplified fragments as follows: products of 68bp, 281bp, 122bp, 119bp, 167bp and 97bp in size belonged to genotype A, B, C, D, E and F, respectively. A sample was defined as a mixed genotype when its products consisted of any two fragments above (Fig. 1).
-
The method for detecting subgenotypes of HBV genotype B was slightly modified according to protocol described elsewhere (7), and it could distinguish between subgenotype Ba and Bj of HBV genotype B. HBV DNA sequences bearing the polymerase region were amplified by PCR with seminested primers. The first-round PCR was performed with a sense primer (PC1: 5'-CATGCAACTT TTTC ACCTCTGCCT-3', nt1813-1836) and an antisense primer (COR: 5'-GAGTGCGAATCCACACT CCA-3', nt2285-2266). The second-round PCR was carried out with another sense primer (PC2: 5'-TGTTCAAGCCT CCAAGCTGTG-3', nt1861-1881) and COR. The two PCR cycles was performed within the same reaction conditions: denaturing at 94℃ for 5 min, then 30 cycles of amplification at 94℃ for 40 s, 55℃ for 30 s, and 72℃ for 50 s, the final extension at 72℃ for 7 min. A portion (5μL) of the second-round PCR product of 425bp in size was digested with 5 U of Stu Ⅰ at 65℃ for 1~2 h and Hpa Ⅰ at 37℃ for 3 h each.
The primers for testing subgenotypes of HBV genotype C were designed according to Tanaka's study (8). The method consisted of two PCR cycles with semi-nested primers followed by RFLP with the restriction site specific for subgenotype Cs or Ce. The primers of the first-round PCR were 964F (sense, 5'-ATTAGACCTATTGATTGGAAAGT-3', nt964-986) and 1272R (antisense, 5'-AGTATGGATCGGCAGAG GAG-3', nt1272-1253). The primers of the secondround PCR were 970F (sense, 5'-CCTATTGA TTGG AAAGTATGTCA-3', nt 970-992) and 1272R. The two PCR reaction conditions were performed under the same conditions as in the PCR for identification of subgenotype Ba/Bj. The second-round PCR products of 309bp in size was first digested with BstEⅡ at 37℃ for 3 h, and those not cut by BstEⅡ were digested with Bcn Ⅰ at 37℃ for 3 h.
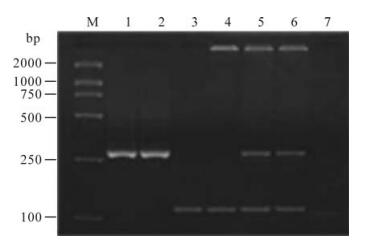
HBV subgenotypes Ba/Bj and Cs/Ce were identified by the size of digested fragments with restriction endonucleases (7, 8). Fig. 2 shows the electrophoresis pattern of the second-round PCR products. Fig. 3 illustrates the electrophoresis pattern of the products with endonuclease digestion.
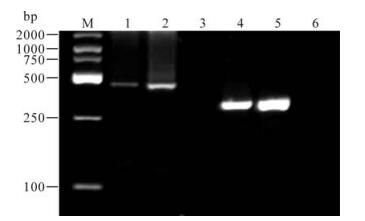
Figure 2. The electrophoresis pattern of HBV subgenotyping products by hemi-nested PCR. M, DNA Marker; 1/2, SecondPCR products of amplifing genotype B (425bp); 3, Negative contral for genotype B; 4/5, Second-PCR products of amplifing genotype C (309bp); 6, Negative contral for genotype C.
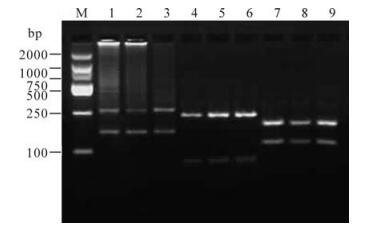
Figure 3. RFLP pattern of identification of HBV subgenotypes. M, DNA Marker; 1-3, subgenotype Ba, two fragments of 265 bp and 160 bp by digestion with StuⅠ; 4-6, subgenotype Cs, two fragments of 233 bp and 76 bp by digestion with BstEⅡ; 7-9, subgenotype Ce, two fragments of 192 bp and 117 bp by digestion with BcnⅠ.
-
Chi-square test was used as appropriate. Differences were considered significant for P values of < 0.05.
Patients
Main reagents
HBV DNA extraction
Genotype-specific primers-PCR for identification of HBV genotypes
PCR-RFLP for identification of HBV subgenotype Ba/Bj and Cs/Ce
Statistical analysis
-
The study indicated that among the 100 patients with chronic HBV-infection, the proportions of genotypes B, C and B+C were 41% (41/100), 25% (25/100) and 34% (34/100), respectively. In this study, genotypes A, D, E and F weren't examined. The relationship between the clinical data of 100 patients with chronic HBV infection and HBV genotypes is illustrated in Table 2. Genotype distribution showed no statistical difference between male and female of patients (χ2=3.406, P=0.182), and among the age groups of patients (χ2=1.585, P=0.812) (Table 3). There were no significant differences in the distribution of HBeAg positive rates among genotype B, genotype C and mixed genotype (B+C) (χ2=5.118, P=0.077). Sinilarly, the positive rates of HBeAb among genotype B, genotype C and mixed genotype (B+C) were not significantly different (χ2=0.823, P=0.663).

Table 2. Relationship between HBV infection and HBV genotypes in 100 patients with chronic hepatitis B

Table 3. The distribution of HBVgenotypes in age groups
-
In the 41 patients with genotype B infection, (100%) all of them infected with subgenotype Ba and none was found to infect with subgenotype Bj. Of the 25 patients with genotype C infection, 21(84%) and 3 (12%) belonged to subgenotype Cs and Ce, respectively, while the remaining 1 that was not identified by PCR-RLFP was further analysed by sequencing.
The distribution of HBV genotypes and its clinical significance
The distribution of HBV subgenotypes
-
HBV genotypes have distinct geographical distributions. Up to now, four genotypes (A, B, C and D) have been reported in China, of which genotype B and C are highly prevalent while genotype A and D play minor roles. Genotype C makes up a major proportion in the northern areas of Mainland China and genotype B increases gradually from north to south where it can be the main genotype (10). This study demonstrated that genotypes B, B+C, and C existed among the Bai nationality in Dali, and genotype B was the predominant genotype. This result corresponds with finding in related domestic reports. The most prominent difference was the higher prevalent rate (34%) of mixed genotype (B+C) in patients, which seems to be specific to the region. Dali is an ethnic minorities inhabited district in which the population of Bai nationality accounts for the main proportion. The construction of population is very complex and people have different life customs in the district but can influence each other, which may be the major cause of the presence of mixed genotype infection. Another possible reason is that the method of genotypespecific primers is fairly sensitive to the examination of mixed genotype infection.
Currently, there is still not a consensus opinion about the relationship between a HBV genotype and its clinical pathogenicity on account of the different HBV prevalent strains in different areas, the sample number, and the genotyping methods. Some studies indicated that patients with HBV infection of genotype B had a lower HBeAg positive rate and an earlier HBeAg seroconversion rate, compared those with genotype C (3, 10). But others argued that there was no association between the rate of HBeAg positivity and genotypes B & C (4). Our study demonstrated that the rates of HBeAg/HBeAg positivity were no significantly different among genotypes B, genotype C and mixed genotype (B+C). Generally speaking, the appearance of HBeAg indicates that replication of HBV is active, and HBeAg seroconversion predicts that the condition of patients will improve. But some studies suggested that HBeAg in some patients with severe liver damage was negative due to the mutations of HBV gene, such as nt1896 mutation in the Pre-C region and a A1762T/ G1764A mutation in the BCP region that can affect the expression of HBeAg (2, 6). Therefore, investigating the epidemic status of HBV mutations in differrent genotypes and expanding the sample size may help to clarify the problem.
The situation regarding the prevalence of HBV subgenotypes in China is still not clear at present. Some investigations showed that subgenotype Ba was the predominant strain in the Mainland of China, and subgenotype Cs was mostly found in the southern part of China while subgenotype Ce mostly in the northern areas (1, 11). Our investigation suggested that among Bai nationality in Dali, all genotype B strains belonged to subgenotype Ba while subgenotype Bj was not found, and for genotype C, 84% were subgenotype Cs, 12% were subgenotype Ce. To some extent, the study demonstrated that subgenotypes Ba and Cs were the predominant strains in patients with chronic HBV infection among Bai nationality in this district. Meanwhile, further studies with an expanded sample size are needed to explore the relationship between the HBV subgenotypes and clinical disease.
In conclusion, this present study reports for the first time the distribution of HBV genotypes & subgenotypes among the Bai nationality in Dali. In addition, the study enriched the data of HBV genotypes & subgenotypes in ethnic minority population and further confirmed previous reports regarding the fact that HBV genotypes & subgenotypes have distinct geographical distributions.













 DownLoad:
DownLoad: