-
Porcine reproductive and respiratory syndrome (PRRS) is considered one of the most economically important diseases affecting the swine industry world-wide, characterized by reproductive failure in late term gestation in sows and respiratory disease in pigs of all ages (25). Porcine reproductive and respiratory syndrome virus (PRRSV), the causative agent, has been classified, together with lactate dehydrogenase elevating virus (LDV) of mice, equine arteritis virus (EAV) and simian haemorrhagic fever virus (SHFV), into the family Arteriviridae in the order Nidovirales (8).
PRRSV, a member of the small enveloped viruses, has a single-stranded, non-segmented, positive-sense, polyadenylated RNA of approximately 15 kb in length that contains nine open reading frames (ORFs). ORF1a and ORF1b encode viral replicase polyproteins, while ORF2a, ORF2b, and ORFs 3-7 encode the viral structural proteins GP2, E, GP3, GP4, GP5, M, and N, respectively (10, 21, 23, 24, 26, 39). Sequence com-parison of PRRSV isolates from different geogra-phical regions indicated that there are two major genotypes represented by prototype viruses Lelystad and VR-2332 with differences in nucleotide sequence of approximately 40%. These viruses are referred to as the European strains (EU, or type 1 [Lelystad virus, LV]) (24) and the North American strains (NA or type 2 [VR-2332]) (27).
However, there is increasing evidence which in-dicates that these PRRSV strains are biologically, antigenically and genetically heterogeneous. They differ in virulence and are associated with different clinical signs (4). Studies have shown that ORF5 and the non-structural protein 2 (NSP2)-coding gene (nsp2) may represent the most genetically variable regions in the PRRSV genome (2, 9, 14, 19, 20, 32). It is also well documented that PRRSV strains differ greatly in their pathogenicity (5, 12, 35).
In this study, we investigated some clinical samples in Hubei province of China and isolated some genes of one emerging PRRSV variant, ZB, which was the causative agent of so-called "high fever" disease in Hubei province in 2006 similar with those PRRSV variants isolated from other provinces of China in the same period. Phylogenetic analyses of three genes, N, GP5 and NSP2, were carried out with the aim of molecular characterization of this particular PRSSV. The ZB-N is identical to the N gene from all the Chinese isolates belonging to North Amercian (NA) type. However, several mutations in GP5 and a novel deletion in NSP2 gene distinct from all of the other previous PRRSV strains in ZB were identified as same as the other PRRSV variants isolated in China in 2006, and considered to be the causes of the change in virulence of this PRRSV variant possibly.
HTML
-
Clinical samples (lungs, livers, kidneys, spleen and lymph nodes) were collected from clinically ill pigs from different pig farms in Hubei province in China during the period from June 2006 to April 2007.
-
To visualize the suspected causative agent, PRRSV, the infected tissues were firstly fixed in 3% glutaral-dehyde, then fixed in 1% osmic acid, and embedded in Epon812. Ultra-thin sections (500-800nm) were pre-pared, stained with uranyl acetate and lead citrate, and examined using EM.
-
PRRSV was detected by reverse transcriptase poly-merase chain reaction (RT-PCR). Total RNA was extracted directly from different tissues (such as lungs, livers, kidneys, spleen, intestine and lymph nodes) using TRIZOL (Invitrogen). Reverse transcription was performed at 37 ℃ for 10 min with 13μL total RNA, 1μL random primer (Promega), 1μL of 10mmol/L dNTPs and 4μL 5×RT buffer, then chilled on ice for 5 min. 1μL moloney murine leukaemia virus (MLV, Promega) RT was added and incubated at 37 ℃ for 1h.
The cDNA was amplified by RT-PCR with the following primers: NF (5'-TAACAACGGCAARCA GCRRAA-3') and NR (5'-TGRYGCTGTGACRCGR ATC-3'), NSP2F (5'-CCTWCTTTGCTCCCCCTTG AATGT-3') and NSP2R (5'-AAGACTTGTTGCGCC ACGGAGGTA-3'), GP5F (5'-atgttggggaagt gcttgaccg-3') and GP5R (5'-ctagagacgac cccatWgttcc-3') using KOD plus Taq DNA polymerase (TOYOBO). The expected amplified frag-ment size was 352 base pairs (bp), 399bp and 603bp, respectively.
The PCR consisted of 35 cycles of denaturation at 94 ℃ for 45s, annealing at 55 ℃ (54 ℃ and 57 ℃) for 45s and extension at 72 ℃ for 1min, with a final extension at 72 ℃ for 10 min. The amplified PCR products were separated by gel electrophoresis.
The complete NSP2 gene was amplified using PCR-driven overlap extension using the following the primers (18): NSP2-1F: 5'-gctggaaagagagcaaggaa ag-3'; NSP2-1R: 5'-GAAACTCTCTCAAGTTTGG CCA-3'; NSP2-2F: 5'-ccaaccgggctactccgg aaga-3'; NSP2-2R: 5'-GGTGTCATCGGCTCGGA TGGTG-3'; NSP2-3F: 5'-ggttttgatgggcgac aatgtc-3'; NSP2-3R: 5'-CACTGAACCAATGGT GAGATCA-3'; NSP2-4F: 5'-aagatgattctcga gacaccgc-3'; NSP2-4R: 5'-GCCCARTAACCTG CCAAGAATG-3'.
After agarose gel electrophoresis, PCR products were excised, purified using Gel Extraction Kit (Omega), and cloned into T-easy vector (Promega). Recombinant clones were sequenced at Invitrogen company with an Automated DNA Sequencer. Sequence results were analyzed using the DNAStar software. The unrooted trees were constructed from the aligned amino acid sequences of all of the PRRSV isolates available in GenBank by distance-based method using the MEGA softwear package (Version 4.0).
Clinical Samples
Electron Microscopy
Detection of PRRSV in different clinical samples with RT-PCR and sequence analysis
-
Since June, 2006, a previously unknown severe disease designated 'high fever' spread throughout more than 10 provinces (autonomous regions), in-cluding Hubei. The representative sick pigs had the following common clinical symptoms: rubefaction, blood spots, petechiae, erythematous blanching rashes, and pimples, frequently observed in the ears, mouth, noses, back, and the inner thigh as shown in Fig 1a. While differing greatly from the general "blue ear" disease, other common symptoms included high fever (40-42 ℃), depression, anorexia, cough, asthma, la-meness, shivering, disorder in the respiratory tract, and diarrhea were observed. To gain insights into the pathological changes of those representative death cases, autopsies were performed on representative mortalities. These autopsies results demonstrated severe damage of multiple organs such as foci with patho-logical changes and hyperplasia in the lungs together with lung haemorrhagic spots and lung edema (Fig. 1C), as well as frequent blood spots in kidney (Fig. 1D). Furthermore, PRRSV like viral particles were observed in infected tissues by electron microscopy (Fig. 1D).
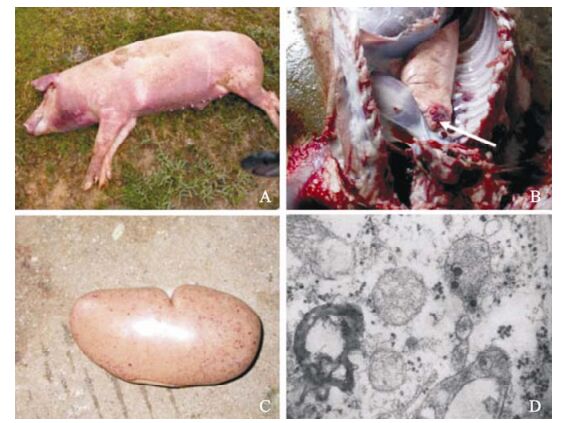
Figure 1. Clinical presentation of pigs with "high fever" disease and the severely damaged organs in dead pigs and the morphogenesis of PRRSV in infected tissue under the electron microscopy (EM). A: The killed pig with red discolorations and pimples in this epidemic. B: Lung haemorrhage (indicated at the arrow). C: Kidney with frequent blood spots. D: The PRRSV particles (indicated by arrows) are visualized inside the mitochondria in ultra-thin section.
More than 100 clinical samples (from serum, lungs, kidneys, liver, and lymph nodes) were collected and 70 samples were found to be PRRSV positive by RT-PCR based on open reading frame 5 (GP5) and N (Fig. 2). Sequence analysis revealed that ORF5 and N from these samples were highly homologous to HB-1, p129 and other North American type strains (data not shown). Viral RNA was detected in many organs and tissues, including liver, spleen, kidney, lung and lymph nodes but not the intestine (Fig. 2).
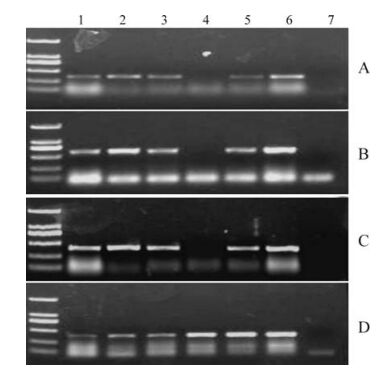
Figure 2. RT-PCR detection of the viral pathogen from the infected tissues with specific primers for PRRSV. A: PCR assay with N primer. B: with GP5 primer. C: with NSP2 primer. D: with actin primer. Lane 1 to 6: the supernatants from the homogenized tissues of liver, spleen, kidney, intestine, lung and lymph, respectively. Viral RNA is present in all in examined tissues with the exception of the intestine (lane 4).
-
To clarify the genetic backgrounds of these isolates, complete N gene (ZB isolate, ZB-N) and GP5 (603bp) gene were directly amplified from infected lung tissues by RT-PCR, and then cloned and sequenced. The N gene of the ZB isolate was 372bp. Phylogenetic analysis of the N gene from this isolate together with the N gene from more than 280 other isolates in GenBank showed that ZB-N is identical to the N gene from all the Chinese isolates belonging to North Amercian (NA) type, the large cluster in the upper part of our estimated tree (Fig. 3A). It shares 95% identity with ATCC VR-2332 (the NA-type proto-type), but only 59% identity with LV and EuroPRRSV (the EU-type prototype). Similar results were also obtained when the GP5 and NSP2 sequences were subjected to phylogenetic analysis. In addition, ZB-N was found to have 100% identity with JXA1, HEB1, HUB2 and the other viruses isolated during the 2006 PRRSV outbreak. These results indicate that ZB is another highly pathogenetic PRRSV variant in circulating China.

Figure 3. Phylogenetic analyses of PRRSV three genes. Three unrooted tree were constructed from the aligned amino acid sequences of all of the PRRSV isolates available in GenBank by distance-based method using the MEGA softwear package (Version 4.0). Bootstrap values were calculated from 1000 replicates of the alignment. Some standard isolates are indicated by different color arrows. A: Phylogenetic analysis of the N gene sequence. B: Phylogenetic analysis of the GP5 gene sequence. C: Deduced amino acid sequence of ZB NSP2 aligned with that of reference strain ATCC VR-2332, and other Chinese strains isolated before 2006 and some isolated during 2006 "high fever" disease outbreak. Amino acids deletions relative to ATCC VR-2332 are indicated by dashes (-). D: Phylogenetic analysis of the NSP2 gene sequence.
The 603bp GP5 gene of the ZB isolate, as a putative virulence marker, was also verified by evolutionary analysis of amino acid sequence. At the amino acid level, there are 88%-94% identities at the GP5 gene between the ZB strain and other NA-type viruses such as ATCC VR-2332 and some typical NA-type Chinese isolates HB-1 (sh)/2002, HB-2 (sh)/2002, and CH-1a. An even higher degree of homology (99%) was found for the GP5 gene between the ZB strain and the recently reported JXA1, HEB1, and HUB2 strains. On the other hand, only 57%-58% identities were found at the GP5 gene between the ZB strain and EuroPRRSV and LV. The phylogenetic tree com-prising more than 400 sequences and the ZB isolate revealed that ZB-GP5 is associated with the NA-type, but localized to another clade of the NA-type cluster and clearly distinct from the clade containing ATCC VR-2332 (Fig. 3B).
The NSP2 gene of ZB isolate amplified using PCR-driven overlap extension was found to be 2850 bp long encoding for 950 amino acids (18). When compared with NA-type prototype, ATCC VR-2332, two deletions were found in the NSP2 gene (1 amino acid at position 480 and 29 amino acids at 520-530) (Fig. 3C). Alignment showed that ZB-NSP2 had more than 97% identity with some newly emergent variants isolated in China during 2006, such as JXA1 and HEB1, 74%-88% identities with other Chinese strains, such as HB-2 (sh)/2002 and HB-1 (sh)/2002. However, it was found that the NSP2 gene of ZB virus shares low level of homology with other PRRSV strains, for example there is only 65% identity with ATCC VR-2332 and approximately 35% identity with Euro-PRRSV. These results indicate high diversity among the NSP2 gene (Fig. 3D). In addition, ZB-NSP2 had different deletions of amino acids at different sites when compared to the low pathogenic Chinese strain HB-2 (sh)/2002 (Fig. 3C).
Phylogenetic analysis was also performed on the NSP2 gene of the ZB isolate and 92 other PRRSV isolate worldwide. ZB-NSP2 clustered within the NA-type together with other 68 isolates of which most were Chinese isolates from different provinces and years and the prototypic North American isolate ATCC VR-2332. Clearly there are two subgroups within this lineage. Most of the Chinese isolates were classified in one subgroup and the rest of isolates from USA and other Asian countries were in the other subgroup (Fig 3D).
Pathology of clinical sample and identification of the causative agent
Sequencing and phylogenetic analysis
-
Porcine reproductive and respiratory syndrome (PRRS) has been recognized as a serious swine disease and is characterized with either reproductive failure in pregnant sows, or respiratory tract distress particularly in sucking pigs (33, 37). To date, PRRS has spread worldwide with the characteristics of an endemic in swine-cultivating countries, causing enor-mous economic losses each year (1, 5, 7, 11).
Phylogenetic analysis of PRRSV isolates from different geographical regions worldwide clearly indicates the existence of two major genotypes: Type Ⅰ representing the European prototype (EuroPRRSV), and Type Ⅱ with the Northern American strain ATCC VR2332 as a prototype (26, 28). In this study, a investigation, ranging from pathology to molecular diagnosis, was carried out with the aim of under-standing the molecular characteristics of a PRSSV variant isolated from Hubei, China in 2006. Phy-logenetic analysis indicate that our isolate, ZB, belongs to the NA-type and is another highly patho-genetic PRRSV variant, and the most of the variants found in 2006 epizootic outbreak of pig diseases in China, including ZB isolate, were the farthest variants from the typical NA-type PRRSV in phylogenetic distance
Initially'high fever' was used to describe the disease in pigs because the symptoms were different from the typical PRRS. Other pathogens were sus-pected until the EM observation of PRRSV particles in the ultra-thin section of infected tissues (Fig. 1) and the detection of PRRSV-specific sequences in various swine tissues by PCR-based assays (Fig. 2). Due to the magnitude of the outbreak and the consequent economic and social impacts it is important to understand how this virulent variant of PRRSV emerged in China and caused so much damage. It has been questioned whether a virulent strain of PRRSV, or novel virulence factors have been acquired by PRRSV ancestors during evolution under some local selection pressures (34, 29, 31). Prior to this outbreak of PRRS, Northern American (VR2332)-like PRRSVs including HB-1, HB-2, and BJ-4 strains existed in China (16). However, there is no evidence to suggest that these viruses were associated with large scale epidemics of PRRS with fatal cases among grown sows (9, 16). It was thought that mixed infection with other pathogens, particularly bacteria could be one of the significant causes. Studies have indicated that intratracheal administration of bacterial lipopoly-saccharides (LPSs) to PRRSV-infected pigs results in markedly enhanced respiratory disease, whereas the inoculation of each component separately results in largely subclinical disease. Gucht and his colleagues (17) had examined whether PRRSV-LPS-induced respiratory disease is associated with the excessive production of proinflammatory cytokines in lungs. Research results revealed that all pigs inoculated with PRRSV-LPS developed severe respiratory disease, whereas pigs inoculated with PRRSV or LPS indivi-dually did not. PRRSV infection significantly en-hanced cytokine production in response to LPS. In particular, TNF-α, IL-1, and IL-6 levels were found to be 10 to 100 times higher in the PRRSV-LPS-inoculated pigs than in the pigs inoculated with PRRSV or LPS alone, and the cytokine level correlated with the severity of respiratory symptoms. Surprisingly, the level of neutrophil infiltration and the pathological changes detected in the lungs of PRRSV-LPS-inoculated pigs resembled those detected when the effects of PRRSV and LPS inoculated individually are combined, but with no synergistic effects between PRRSV and LPS. The reason for these observations is still unclear, but these data demonstrate a synergism between PRRSV and LPS in the induction of proinflammatory cytokins and an association between induction of these cytokines and disease. Additionally, the autopsy results from dead pigs which died in the 2006 outbreak also demon-strated severe damage of multiple organs such as the lungs (Fig. 1).
Undoubtedly, the Northern American-like PPRSV virulent strains found to be the etiological agents for this epidemic was another important reason. The sequence analysis showed extensive amino acid (aa) mutations in the GP5 protein and deletions in Nsp2 (1 aa deletion at 481, and 29 aa deletion at 520–530) in all the isolates associated with the "high fever" disease outbreak in 2006 (Fig. 3C) when compared with the sequence from previous Chinese isolates CH-1a and HB-1 (sh)/2002. These evidences imply that the novel molecular hallmarks of these PRRSV variants may correlate with their high pathogenicity.
GP5 is a very important structure protein in PRRSV and is responsible for attachment of the virus to the host cell and contains the primary neutralization epitopes involved in virus neutralisation (3, 38). Extensive genetic variation has been observed among PRRSV isolates and the observed genetic hetero-geneity of PRRSV could lead to the selection of a more virulent virus and the consequence emergence of new forms of PRRSV. As an example, an acute strain of PRRSV has caused high abortion and mortality in vaccinated swine populations in the USA (6). Sequence analysis showed that these acute PRRSV isolates shared 88–96% amino acid sequence identities with each other or with Northern American PRRSV isolates and the modified live vaccine (RespPRRS MLV) (19). In the present study, the GP5 genes of ZB isolates shared 99% predicted amino acid sequence identity with those isolates from herds of pigs experiencing severe disease outbreaks in 2006, but only 87-88% amino acid sequence identity with RespPRRS MLV vaccine strain and NA-type prototype, ATCC VR-2332 (Fig. 3B). These findings could explain why vaccination with the current vaccine failed to prevent the emerging epidemic caused by the PRRSV variant.
Nsp2 is a multidomain protein and has been shown to undergo extensive remarkable genetic variation, primarily in its middle region. In our study, phylo-genetic analysis showed that the ZB-Nsp2 gene was clustered with the NA-type, but shared only 65% predicted amino acid sequence with identities with prototypic ATCC VR-2332 and 81% identity with those of Chinese PRRSV isolates from previous outbreaks. Pairwise homology comparison of deduced amino acid sequences seemed to support the notion of increased genetic distances between early and current isolates. More importantly, there are deletions of one and 30 amino acids relative to the Nsp2 protein of VR-2332 which was different from most of the previous Chinese isolates though amino acid deletions in Nsp2 which have been identified in some PRRSV isolates (16). Recently, many research groups reported that areas in Nsp2 with deletions and amino acid hypervariability are predicted to be likewise immuno-logically important (15), and Nsp2 might function as an immunodominant protein under selective pressure by host immune responses (30). It is thus possible that NSP2 gene with this unique molecular hallmark is responsible for the "high fever" disease outbreak observed in 2006. Further experimentation is required to validate this hypothesis.
Based on all of the GP5 and NSP2 sequences available in GenBank, the inferred phylogenic trees revealed a high degree of variability among the NA-type PRRSV, especially the Chinese isolates. It is generally believed that strain evolution will result in an ever-broadening strain diversity for both genotypes and it is difficult to predict what impact this will have on the clinical picture and the immune response of the host (13, 22).
In summary, the ZB isolate from Hubei province China in 2006 belongs to the NA-type, and is another highly pathogenic PRRSV variant. Based on the GP5 and NSP2 phylogenic analysis, a high degree of variability is apparent between the high pathogenic PRRSV variants isolated from "high fever" disease outbreak in China in 2006 and the other previous Chinese isolates and prototypic NA-type PRRSV. It is likely that these differences manifest themselves as differences in pathogenicity and effect on the host immune system, and this is currently under investi-gation in our laboratory.













 DownLoad:
DownLoad: