-
Recent studies have revealed that a number of insect baculoviruses (Baculoviridae) exist as a mixture of genotypes that can simultaneously infect their hosts (5, 10, 12, 26). Baculoviruses are ideal model organisms for studies on host-virus interactions because they often naturally survive as mixed-genotype populations, they are highly amenable to laboratory manipulation, both in insect cell lines and in insects, a great deal of genomic sequence information is available in data-bases, they are easily quantified and have an out-standing safety record.
The genome of these arthropod pathogenic viruses is a circular, double-stranded DNA molecule that is included in a protein capsid, constituting the nucleoca-psid. Nucleocapsids are enveloped by a lipoprotein membrane to form the infectious particles or virions. Outside their hosts, baculoviruses are found as large proteinaceous occlusion bodies (OBs) in which one or more virions are embedded. The usual route of infection is the ingestion of OBs by susceptible larvae. OBs dissolve in the alkaline environment of the host midgut, releasing enzymes and occlusion derived virions (ODVs) that cross the peritrophic matrix and initiate a characteristic biphasic infection cycle. The ODVs set off the primary infection by fusion with the columnar epithelial cells of the larvae. This first step is enabled by the presence of a group of viral encoded proteins such as P74 (16, 30, 32), PIF1 (14), PIF2 (23), PIF3 (21) and PIF4 (Ac150) (17, 31). Infected cells give rise to a second morphotype, the budded virus (BVs), which are responsible for disseminating a systemic infection within the host.
Lepidopteran nucleopolyhedroviruses (NPVs) (genus Alphabaculovirus) are characterized by having multiple virions occluded within each OB. Commonly observed features of NPVs are the coexistence of multiple genot-ypes within the same virus isolate (5, 15, 18). This genetic diversity can be observed at the level of single larvae (4, 5, 22). It has been demonstrated that in larvae, individual cells are usually infected by mul-tiple BVs (2, 9, 24), and the genotypes in these BVs might be diverse. The importance of such diversity in the insecticidal properties of the virus population can be studied using experimental mixtures of genotypes, in addition to the observation of the behavior of natural virus populations.
In a Nicaraguan isolate of Spodoptera frugiperda multiple nucleopolyhedrovirus (named SfNIC), eight out of nine plaque purified variants were identified as genotypes that contained deletions (named SfNIC-A, and SfNIC-C to SfNIC-I). These deletions represented between 4 and 11% of the genome compared to the genome of the single complete genotype, named SfNIC-B (26). SfNIC-C possess the largest deletion (16.4 kb) that affects a number of genes including pif1 and pif2. For this reason it is unable to infect larvae per os (26). However, this genotype can survive by complementation with the complete genotype (SfNIC-B). In cells that are simultaneously coinfected by both genotypes, both PIF1 and PIF2 proteins are produced by the complete genotype rendering the ODVs in-fectious per os, irrespective of the genotypic com-position of the progeny nucleocapsids. Moreover, remarkable cooperative behavior has been demons-trated to occur in mixtures comprising SfNIC-B and SfNIC-C. The LC50 value of OBs comprising SfNIC-B is ~2.5-fold higher than that of wild-type SfNIC OBs, indicating reduced potency of SfNIC-B OBs. How-ever, when ODVs were released from mixtures com-prising ~75% of SfNIC-B OBs and ~25% of SfNIC-C OBs, and injected into insects that subsequently died of polyhedrosis disease, the resulting OBs comprising co-occluded genotype mixtures had an insecticidal potency similar to that of the wild-type isolate (19). Recent studies with pif1/pif2 deletion recombinants have revealed that (ⅰ) it is the deletion of this region that is responsible for the observed potentiation of insecticidal potency in mixtures of complete and deletion genotypes and, (ⅱ) the proportions of com-plete and deletion genotypes present in mixtures subjected to serial passage per os rapidly converge to a genotype composition that results in the highest virus transmissibility (3).
Whether this selection for specific ratios of genotypes occurs mainly during the establishment of the primary infection process in insect midgut epithelial cells or during the later development of the systemic infection of the host is unknown. Recently, Zwart et al. (33) attributed negative selection for defective genotypes to changes in the average multiplicity of infection (MOI) during the systemic infection process. In an attempt to elucidate this question, we analyzed the frequency of SfNIC-B and SfNIC-C genotypes at different time points throughout the infection process in larvae inoculated with different co-occluded mixtures of these two genotypes.
HTML
-
Spodoptera frugiperda larvae are a laboratory colony maintained at 25℃, 75% RH, and 16 h light: 8 h dark photoperiod on a semi-synthetic diet (7). Two distinctive genotypes, SfNIC-B and SfNIC-C, were used in this study. These genotypes were purified in vitro from a natural population of S. frugiperda nucleopolyhedrovirus (SfMNPV) originally isolated in Nicaragua (SfNIC) (6). SfNIC-B genotype was the only genotype with a complete genome; all other genotypes presented deletions of varying length in one region of the genome (26). The SfNIC-C genotype presents a 16 kb deletion and is not infectious per os but is capable of replication following injection or when co-infected with a genotype that is infectious per os. OBs of SfNIC (wild-type), SfNIC-B and SfNIC-C were amplified by droplet-feeding (13) fourth instar larvae in the case of SfNIC and SfNIC-B or by intra-hemocelic injection in the case of SfNIC-C. OBs were collected from dead diseased insects, filtered through muslin and centrifuged to eliminate insect debris. OB concentrations were determined using an improved Neubauer hemocytometer (Hawksley, Lancing, UK) under phase-contrast microscopy. Viral DNA extraction from a suspension containing ~109 OBs was performed following the method described by Simon et al. (27). Restriction endonuclease analysis was used to verify the DNA profiles of both purified genotypes and the wild-type isolate.
-
To obtain co-occluded genotype mixtures, purified OB suspensions of the SfNIC-B and SfNIC-C genot-ypes were diluted to a concentration of 5×108OBs/mL, and dissolved in alkaline buffer (1 vol. OBs: 1 vol. Na2CO3 0.1mol/L: 5 vol. H2O) at 60℃ for 10 min. to release ODVs. Undissolved OBs were pelleted at 2 655×g for 5 min and discarded. Each ODV-containing supernatant was then mixed in one of three different proportions: 90:10, 50:50 and 10:90. These mixtures were named the co-injection samples (INJ), as they were injected into groups of 50 S. frugiperda fourth instars (~8 μL/larva) from the laboratory colony. These larvae were individually reared until death. The OBs obtained from these larvae were purified and pooled, and each sample, named Day zero samples (D0), was used as inoculum to infect second instars per os by the droplet feeding method. Groups of 50 larvae were inoculated with one of the three co-occluded mixtures at a concentration of 1.2×106 OBs/ mL, which was estimated to result in ~90% mortality. Groups of eight larvae were sampled every 24 h during a five day infection period, anesthetized on ice, surface-sterilized with ethanol and their prolegs were cut to obtain the hemolymph, which was transferred im-mediately into ice-cold, sterile eppendorf tubes con-taining 5 μmol/L L-cysteine to prevent melanization. A haemocyte-free suspension was obtained by centrifu-gation at 664×g for 5 min. at 4℃. Supernatants, consti-tuting the samples taken at 1 to 5 days post-infection (D1-D5), were stored at -20℃ until further use. The whole procedure was performed three times.
-
The relative proportions of SfNIC-B and SfNIC-C present in: ⅰ) the ODV mixtures that constituted the co-injected samples (INJ), ⅱ) the D0 samples of OB inocula and, ⅲ) the BVs from hemolymph samples taken at 1 to 5 d post-infection were analyzed by PCR using primers designed by Simon et al. (26) that differentially amplify the two genotypes (Fig. 1). The lengths of the amplified fragments were estimated to be 763 bp for SfNIC-B and 646 bp for SfNIC-C (Table 1). PCR reactions were stopped in the linear phase of amplification (19 cycles) as previously described (19). Each amplification was performed three times and amplicons were then purified using a PCR purification kit (Roche, Sant Cugat del Valles, Spain). PCR products were separated by 1% agarose gel electrophoresis and the relative proportions of individual genotypes estimated by densitometric analysis of the intensities of their respective PCR products using QuantityOne (BioRad, Alcobendas, Spain) image analysis program as described by López-Ferber et al. (19).
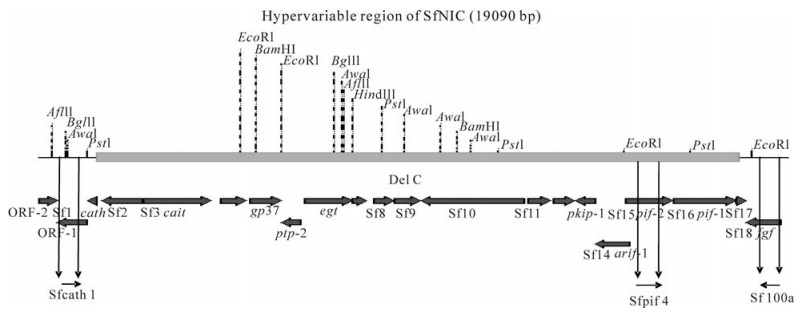
Figure 1. Illustration of the locations of primers used to differentiate SfNIC-B and SfNIC-C genotypes present in mixtures by semi-quantitative PCR within the hypervariable region of the SfNIC genome.

Table 1. PCR primer combinations used for the amplification of SfNIC-B and SfNIC-C and sizes of the products.
Insects and viruses
Experimental virus populations
Quantification of the relative proportions of geno-type mixtures
-
Only slight differences were observed in the relative proportions of SfNIC-B and SfNIC-C present in the primary injected inocula (INJ) compared to the proportions in which each genotype was present in the progeny OBs (D0 samples) that were extracted from injected larvae that died of polyhedrosis disease (Fig. 2). This was so irrespective of the composition of the primary inoculum for all three experimental popu-lations. Similar findings were reported by Simón et al. (24), indicating that the inoculated ratio of these genotypes does not influence their replication rate in the secondary phase of the infection cycle involving systemic infection of the host.

Figure 2. Semi-quantitative PCR of genotypic mixtures produced in insects. Larvae of Spodoptera frugiperda were co-infected with ODVs released from pure genotype OBs that had been mixed at different proportions (ⅰ) 10% SfNIC-B + 90% SfNIC-C, (ⅱ) 50% SfNIC-B + 50% SfNIC-C and (ⅲ) 90% SfNIC-B + 10% SfNIC-C. Mixtures were injected into S. frugiperda larvae (Inj.) and the resulting OBs were used as inocula to infected insects per os (D0). Primers were designed to amplify charac-teristic fragments of SfNIC-B (763 bp) and SfNIC-C (646 bp). PCR reactions were stopped prior to saturation, electrophoresed and densitometric analysis was performed using the intensity of the amplicon generated from SfNIC-B as the reference. Relative density values are shown above or below the corres-ponding amplicon. Molecular markers were M1 1 Kb DNA ladder (Stratgene, La Jolla, CA) and M2 was 100 bp DNA ladder (Invitrogen Corp., Carlsbad, CA) in (B). In both cases, the minority product present following amplifi-cation of the Sf-NIC wild-type is a result of deletion genotypes C and D present in the natural population.
OB samples (D0) were used as inocula for per os infection of S. frugiperda second instars. Over the course of five days of infection, the genotype frequencies differed in each experimental population (Fig. 3). In larvae inoculated with OBs comprising 10% SfNIC-B + 90% SfNIC-C, a dramatic change in genotype fre-quencies occurred between inoculation (D0) and the day 1 sample (Fig. 3A). By day 1, the relative pro-portion of SfNIC-B had increased over five-fold, whereas that of SfNIC-C decreased by two-fold. If we assume that infection of cells within the host occurs at random, insects that are injected with mixtures com-prising a high proportion of SfNIC-C will result in a high proportion of cells being infected exclusively by SfNIC-C genotypes, generating inoculum OBs (D0) that will not contain PIF1 or PIF2 proteins, and these will not be able to infect the midgut. Consequently, each and every OB produced in cells infected by SfNIC-C alone will be cleared in the following per os passage. Only OBs that were produced in cells in-fected by at least one SfNIC-B genome that harbors pif1 and pif2 genes will produce OBs capable of infecting the midgut cells in the following peroral passage. As the protein composition of the OBs reflects the global availability of viral proteins in the nucleus, it is possible that in cells coinfected by SfNIC-C and low numbers of SfNIC-B, OBs may be produced that do not contain any SfNIC-B genomes, but which contain PIF1 and PIF2 proteins in the ODV envelopes, and thus, will be capable of per os transmission. This pseudotyping process has been observed previously when pif1-defective viruses were rescued by transient expression of the pif1 gene carried by a plasmid (8). As a result, the frequency of SfNIC-C in the popu-lation will be quickly reduced. Mathematical modeling predicts that SfNIC-C would disappear unless an advantage for this genotype exists (24). This advan-tage could be, for example, a faster rate of replication due to its slightly shorter genome. If SfNIC-C has an advantage during replication we would expect to observe variation in the relative frequencies of each genotype in each infected insect over time.

Figure 3. Dynamics of genotypic composition in mixtures sub-jected to an infection process in insects. Composition of experi-mental mixtures comprising SfNIC-B + SfNIC-C at OB ratios of (A) 10:90, (B) 50:50 and (C) 90:10, respectively. In all cases, OBs produced in insects injected with genotypic mixtures were subjected to subsequent peroral infection. Samples were taken every 24 hrs. and subjected to semiquantitative PCR and densitometric analyses at each day. Vertical bars indicate S.D. of densitometric readings for three replicate reactions per sample.
In the period from 2 to 5 days post-infection, the frequencies of SfNIC-B and SfNIC-C varied less significantly, with SfNIC-B gradually increasing its relative abundance to 65% at 5 days post-infection (Fig 3A). Although smaller, this variation could indicate that competition efficiency for the host res-ources between both genotypes is taking place. It is known that the MOI changes significantly during the infection process. The number of viruses initiating an infection is low (28) while toward the end of the infection, the MOI is quite high (1, 2). These different conditions evidently cause different selection pressures on both genotypes during the course of infection rather than just the virus entrance suggesting that not only a predominant selective pressure is acting but also a genotypes ability to autonomously replicate and compete for resources is taking place.
In larvae fed with inoculum derived from equal proportions of OBs of both genotypes, no major changes were observed during the period of infection, and the final genotype ratios were 57% SfNIC-B + 43% SfNIC-C at 5 days post-infection (Fig. 3B). In this situation, it is likely that a high proportion of the inoculum (D0) OBs were produced in cells coinfected by both genotypes. This is reflected in the insignifi-cant change in genotype relative proportions between the inoculum OBs (D0) and samples of hemolymph taken at day 1 post-infection.
Finally, in the experimental population consisting of 90% SfNIC-B + 10% SfNIC-C (Fig. 3C), frequencies of genotypes were observed to reach 87% SfNIC-B and 3% SfNIC-C in samples taken at 1 and 2 days post-infection but this proportion changed quite signifi-cantly by day 3, when the relative proportion of SfNIC-C increased to 20% and stabilized at this level during days 4 and 5 post-infection.
It is clear that the lack of infectivity of SfNIC-C OBs is caused by the deletion of pif1/pif2 genes, resulting in ODVs that lack PIF proteins and which are not infectious per os (8, 21, 23, 29). However, the infectivity of SfNIC-C OBs can be restored if the gene is provided in trans, by a plasmid (14), or by the simultaneous presence of a complete genotype in the infected cell (19, 24). The global behavior observed in the SfNIC-B and SfNIC-C mixed genotype OB popu-lations can thus be explained by a combination of two different mechanisms: the midgut barrier that blocks all the ODVs with no functional PIF-1 and PIF-2, and a positive selection of the SfNIC-C viruses, at least when the frequency of SfNIC-C is extremely low, reflected in the increase of SfNIC-C over the duration of the infection (Fig. 3C). The forces driving this advantage may be related to the previous finding that deletion of pif1 increases cell viability and virus driven protein expression (8). The final frequency of SfNIC-C will rely on the outcome of two different and opposing forces, the impossibility for a cell infected only by SfNIC-C to produce infectious OBs will tend to reduce the proportion of SfNIC-C, whereas the specific replication advantage of this genotype during systemic infection of each insect.
When subjected to serial per os passage, mixed populations of SfNIC-B and SfNIC-C genotypes at different proportions consistently converged to a single stable equilibrium after four passages (24). Impor-tantly, however, the most dramatic changes in popu-lation genotype composition were observed at the very first passage, which like the present study, reflects the instantaneous exclusion of OBs that originated from cells infected exclusively by SfNIC-C during the primary infection of insect midgut cells. Near id-entical results were observed in a subsequent study in which the same experiment was performed using experimental mixtures of SfNIC-B and a recombinant virus deleted in pif1 and pif2 (3). Infections by multiple genotypes have been shown to influence different components of nucleopolyhedrovirus fitness, particularly those related to the transmissibility of the virus (11, 19, 20). Few studies on baculoviruses have demonstrated this as clearly as those involving the complete and defective genotypes present in the Nicaraguan isolate of SfMNPV (19, 24-27). The mechanism by which pif1/pif2 deficient genotypes potentiate the insecticidal potency in mixtures with complete genotype B remains to be elucidated. Nonetheless, here we demonstrate that critical selection for certain proportions of genot-ypes occurs principally during the initial step of ODV infection of the host midgut cells. The equilibrium frequency of deletion genotypes in the population appears to represents a balance between the elimi-nation of PIF deficient ODVs in the insect gut and positive selection for fast-replicating deletion variants that may be favored during the systemic infection phase in each insect. The magnitude of within-host selection is likely modulated by changes in average MOI values during the infection period. Clearly, these are intriguing hypotheses on which to base future research.













 DownLoad:
DownLoad: