-
The tick-borne Crimean-congo hemorrhagic fever virus (CCHFV) is a geographically widespread pathogen that causes severe hemorrhagic fever with high mortality rate [11, 12] in human and animals. CCHFV belongs to the Nairovirus genus of the family Bunyaviridae, which is enzootic and has been observed in the Middle East, Asia, Africa, and has also been found in Southeast Europe[1, 2, 6]. The CCHFV particle consists of the small (S), medium (M), and large (L) RNA segments which encode the viral nucleocapsid (NP), glycoprotein precursor (GP), and polymerase proteins, respectively[10]. In China, the CCHF disease was first discovered in Bachu county of Xinjiang autonomous region in 1965. Subsequently, there have been several local outbreaks in southern Xinjiang before 2003 causing serious infections [5]. Diagnosis and epidemiological studies of CCHFV infections are important for CCHF control in China.
It has been reported that monoclonal antibodies (mAbs) 1E10, 1F1, 2C4, 2E10, 1B7, 1B8, 1E5, and 3B6 recognize NP fragments containing amino acid residues between 201 and 306 and its conformational epitopes[9]. Also this fragment is the main antigenic region of CCHFV NP [8]. In this study, we determined the antigenic region of NP by constructing and the expression of a series of different length NP fragments in E. coli. It was demonstrated that the 235-305 aa region of the NP could be identified by a polyclonal serum (rabbit) and 2 mAbs (14B7 and 43E5) against CCHFV using Western-blot analyses. It is expected that this region contain an important epitope which facilitate the diagnoses and vaccine development against CCHFV.
HTML
-
The anti-CCHFV polyclonal serum was generated in rabbit by inoculation of total proteins from purified CCHFVs. MAbs 14B7 and 43E5 were generated in our laboratory and previous studies have shown that they are specific to NP protein [4]. The purified ascites of MAbs 14B7 and 43E5 were used for detections.
-
In order to map the epitope region, the entire NP gene and a series of trunctated fragments of NP were cloned for expression (Fig. 1). Primers were synthesized according to the sequence of strain YL04057 (GenBank accession no. FJ562093) (Table 1). PCR was performed to amplify the fragments from plasmid pT-S which contains the whole S fragment of strain YL04057 CCHFV (stock in our lab). The clones were sequenced for verifying the veracity of the fragments. Then, the fragments were subcloned into plasmid pGEX-KG or pET-32a. The fusion proteins were expressed in E. coli and SDS-PAGE was performed for detecting fusion protein expressions.
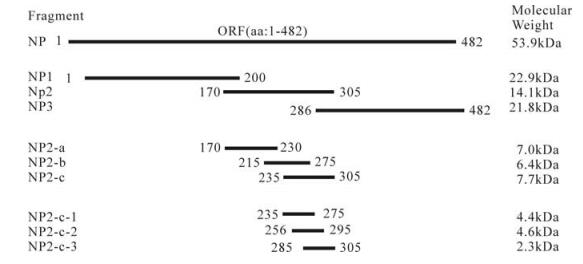
Figure 1. The entire NP and truncated NP fragment of CCHFV expressed in this study. The designations and amino acid positions of the truncated NP fragments are indicated. Red lines marks the fragments which were recognized by the antibodies in Western blot.

Table 1. Primers for clone entire NP and truncated NP fragments of CCHFV
-
E. coli cells expressing each of the NP fragments were resuspended in an appropriate amount of PBS solution and sonicated. After boiling for 10 minutes with sample buffer (0.08 mol/L Tris/HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.005% bromophenol blue) and centrifuged at 12 000×g for 10 min, 5 μL of each supernatant and pellet sample were loaded on a SDS-PAGE gel for electrophoresing. Proteins were stained by using Coomassie brilliant blue R-250 (Sigma, USA).
Equal quantities of samples were loaded onto a 12% SDS-PAGE gel and the gel was electro-transblotted into a nitrocellulose membrane (0.45 μm). The membranes were incubated in TBS containing 5% skimmed milk for 2 h to block nonspecific signals, and then incubated with mAb 14B7 or 43E5 (1:5 000 diluted), or with the anti-CCHFV polyclonal antibody (1:2 000 diluted), and 0.05% Tween-20 (TBST), for 1 h at room temperature. After being washed in TBST, the membrane was incubated with secondary antibodies AP-conjugated goat anti-mouse IgG antibody (1:2 000 diluted) and AP-conjugated goat anti-rabbit IgG (1:2 000 diluted) respectively, for 1 h at room temperature. The positive bands were stained by NBT/BCIP kit (SABC Co.) after washing by TBST buffer.
Antibodies
Expression of truncated NP fragment
Detection of epitopes of antibodies by Western blot analyses
-
The entire NP gene and truncated NP gene were successfully amplified by PCR with designed primers. And four recombinant plasmids pT-np, pT-np1, pT-np2 and pT-np3 were extracted and then confirmed by restriction enzyme digestion (EcoR I and BamH I) and sequencing. After the plasmids were digested, fragments DNA of expected size were obtained, and were inserted into the multiple cloning sites (EcoR I and BamH I) of the pGEX-KG or pET-32a. The recombinant plasmids pGEX-KG-np, pET-32a-np1, pGEX-KG-np2, and pET-32a-np3 were confirmed by restriction enzyme digestion (EcoR I and BamH I) and gel electrophoresis. And other recombinant plasmids pGEX-KG-np2, pGEX-KG-np2-a, pGEX-KG-np2-b, pGEX-KG-np2-c, pGEX-KG-np2-c-1, pGEX-KG-np2-c-2 and pGEX-KG-np2-c-3 were constructed and confirmed according to the same method (Fig. 2).
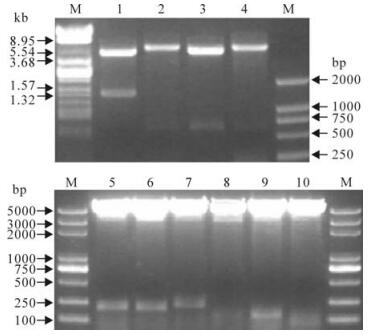
Figure 2. Restriction enzyme analyses of the recombinant expression plasmids. The plasmids were digested with EcoR I and BamH I. 1, pGEX-KG-NP; 2, pET-32a-NP1; 3, pGEX-KG-NP2; 4, pET-32a-NP3; 5, pGEX-KG-NP2-a; 6, pGEX-KG-NP2-b; 7, pGEX-KG-NP2-c; 8, pGEX-KG-NP2-c-1; 9, pGEX-KG-NP2-c-2; 10, pGEX-KG-NP2-c-3; M, DNA ladder.
-
In order to verify the expression of recombinant plasmids, the plasmid DNA of pGEX-KG-np, pGEX-KG-np2, pGEX-KG-np2-a, pGEX-KG-np2-b, pGEX-KG-np2-c, pGEX-KG-np2-c-1, pGEX-KG-np2-c-2 and pGEX-KG-np2-c-3 were transformed into E.coli DH5α cells. The plasmid pET-32a-np1 and pET-32a-np3 were transformed into E.coli BL21 (DE3) cells. The fragments cloned into pGEX-KG vector and the predicted protein were fused with GST with a size about 26kDa; the fragments cloned into pET-32a vector and the predicted protein were fused with Trx tag, S tag and His tag as the size about 18kDa. The SDS-PAGE result showed that relatively large amounts of NP (lane 2, about 78 kDa), NP1 (lane3, about 42kDa), NP2 (lane 4, about 40 kDa), and NP3 (lane 5, about 38 kDa) were expressed in E.coli when induced by 1 mmol/L IPTG, but not so clearly of NP2-a (lane 6, about 33 kDa), NP2-b (lane 7, about 32 kDa), NP2-c (lane 7, about 33 kDa), NP2-c-1 (lane 9, about 30 kDa), NP2-c-2 (lane 10, about 30 kDa) nor NP2-c-3 (lane 11, about 28 kDa) (Fig. 3A).
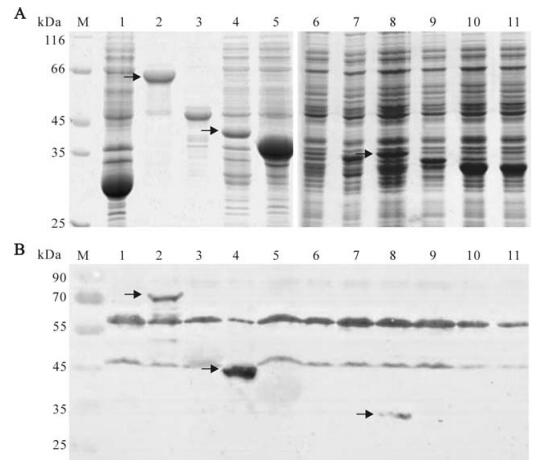
Figure 3. SDS-PAGE (A) and Western-blot (B) analysis of the expression of the entire NP and truncated NP fragments of CCHFV. (A) Crude material from the pellet fraction of the cells transfected with pGEX-KG (lane 1), pGEX-KG-NP (lane2), pET-32a-NP1 (lane 3), pGEX-KG-NP2 (lane 4), pET-32a-NP3 (lane5), pGEX-KG-NP2-a (lane 6), pGEX-KG-NP2-b (lane 7), pGEX-KG-NP2-c (lane 8), pGEX-KG-NP2-c-1 (lane 9), pGEX-KG-NP2-c-2 (lane 10) or pGEX-KG-NP2-c-3 (lane 11) were loaded. M, molecular mass markers. (B) Fusion proteins were detected by Western-blotting with polyclonal anti-CCHFV serum. The three red arrows located positive bands.
-
All recombinant proteins were detected by Western-blot analysis using the polyclonal anti-CCHFV serum; the result showed that NP, NP2 and NP2-c fusion proteins reacted with the anti-CCHFV antibody, but not the other fusion expressed proteins (Fig. 3B). The antigenic region of CCHFV NP was further investigated with mAbs 14B7 and 43E5. The Western-blot analyses showed that both mAb 14B7 and 43E5 could bind to entire NP, NP2 and NP-2-c fragments (Fig. 4). Therefore, we conclude that the region of 235-305aa contains an epitope that can be recognized by polyclonal anti-CCHFV antiserum and mAbs 14B7 and 43E5. This region is a highly conserved region of NP [8].
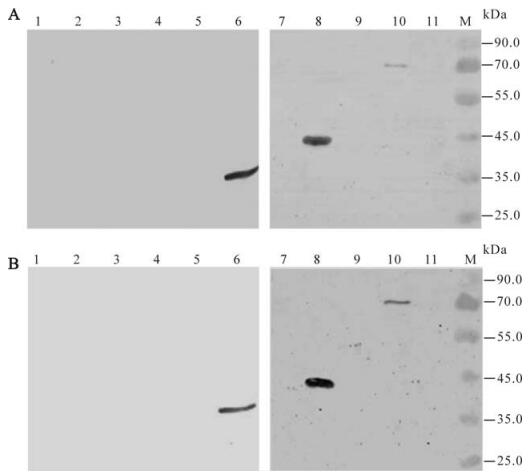
Figure 4. Western blot analyses of the expression of the truncated NP fragments of CCHFV with mAb 14B7 (A) and 43E5 (B), respectively. Fusion proteins NP2-c-1 (lane 1), NP2-c-2 (lane 2), NP2-c-3 (lane 3), NP2-a (lane 4), NP2-b (lane 5), NP2-c (lane 6), NP1 (lane 7), NP2 (lane 8), NP3 (lane9), NP (lane10) and Negative control (lane11) were detected by Western-blotting with mAb 14B7 (A) and 43E5 (B). M, molecular mass markers.
Construction and identification of the recombinant plasmids
Expression of recombinant NP fusion protein in E.coli
Western blot analysis
-
In this study, we cloned the entire NP and truncated NP fragments into prokaryotic expression vectors and the expressed proteins were detected by polyclonal and mAbs for identification of the epitope region. Initially we tried to express all the fragments with pGEX-KG vector, however, it turned out that the np1 and np3 fragments could not be highly expressed in E.coli (DH5α) (data not shown). Finally, we were able to express np1 and np3 with the pET-32a vector in BL21 (DE3) cells. By Western blot using polyclonal and mAbs, we were able to identify an antigenic region at 235-305 aa of the NP protein.
Previously, Saijo et al. (2002) have shown that the CCHF patients' sera with high titers reacted only with the NP fragment containing amino acid residues between 201 and 306 in Western blotting. This is in agreement with our current founding. It can be concluded that the 235-305 aa region of NP protein is an antigenic region in animals as well as in humans.
Previous research on epitope mapping by phage display random gene fragment libraries indicated that specific mAb can recognize epitopes that of 4 or 5 aa [3]. The antigenic regions recognized by mAb 14B7 and 43E5 we identified are 70aa. Therefore, we tried to express smaller peptide as 235-275 aa (np2-c-1), 256-295 aa (np2-c-2) and 285-305 aa (np2-c-3), which were predicted to be potential linear B-cell epitopes [7], in pGEX-KG in E.coli (DH5α). But the expressed fusion proteins were not sensitive to the polyclonal anti-CCHFV serum nor to the mAb 14B7 and 43E5 (Fig. 3, Fig. 4). It might be that the epitope recognized by the 14B7 and 43E5 were disrupted or the predicted location was not accurate, and truncated fragments of NP2-c-1(4.4 kDa), NP2-c-2 (4.6 kDa), NP2-c-3 (2.3 kDa) are too small to expose the potential epitope while fused GST is 26 kDa. So, other methods are necessary to localize the exact epitopes of CCHFV antibodies.
The aa 235-305 region of YL04057 was compared to CCHFV isolates of Chinese strains (Genbank accession no. AJ010648, AJ010649, AF354296, AF358784, AF358596, AF362080, AF415236, AY029157, CHU88413, DQ211642, DQ227495, DQ227496 and DQ217602). The aa 235-305 region of YL04057 were same as DQ227495, DQ227496, DQ217602, AF358784 and AF415236, while the other seven strains were same as each other. There are only two amino acid residues differences among the two groups (data not shown). mAb 14B7 and mAb 43E5 were constructed based on sequence AF354296 but could detect the aa 235-305 region of YL04057. So a difference of two amino acid residues does not affect the result of Western-blot. The identified of a conserved antigenic region will facilitate the development of the CCHFV diagnosis kits as well as provide useful information for vaccine development.













 DownLoad:
DownLoad: