-
Hepatitis B virus (HBV) infection is one of the most prevalent chronic viral infections of humans, with there being approximately 300 million chronic carriers worldwide. Chronic infection can have serious consequences, including cirrhosis and hepatocellular carcinoma, and results in at least one million annual deaths[17, 21, 37]. The most commonly used drugs, interferon-α and nucleoside analogues have only a limited response and none is capable of completely eradicating the virus [10, 18, 24, 27].
Targeted delivery of therapeutic drugs to HBV-infected cells should increase the therapeutic index, reduce the concentration of drug required, and minimize potential side effects [7, 9]. As hepatitis B virus surface antigen (HBsAg) is a common antigen on the surface of HBV-infected hepatocytes, it provides a perfect target for drug delivery [4, 16, 22, 30, 32].
Whereas antibodies have the ability to recognize HBsAg with high specificity and affinity, their large sizes and immunogenicity often limit their pharmacological significance. Humanized antibodies, antibody fragments, and short peptides show great promises but are still restricted by peptidase susceptibility and the immune response. The recent development of the SELEX (systematic evolution of ligands by exponential enrichment) process has provided a new alternative, yielding nuclease-resistant oligonu-cleotides that can be selected to bind tightly and specifically to a given ligand [2, 8, 34]. The diversity of structures exhibited by an aptamer library allows selection of tight binding aptamers for simple targets, such as a single amino acid [6], or for complex targets such as tumor cell lines [11]. Such oligonucleotides, termed "aptamers, " have been made against over 150 different ligands and are emerging as a new class of molecules that contest antibodies in therapeutics, imaging, and diagnostics [12, 13].
Herein, we report the selection of RNA aptamers that can specifically bind to the HBsAg protein. One high affinity aptamer, termed HBs-A22, was isolated from an initial 115 mer library of ~1.1×1015 random-sequence RNA molecules using the SELEX procedure. For further application, the binding activity of aptamer HBs-A22 to HBsAg-positive cells were also evaluated.
HTML
-
The initial RNA library, designated as 25N, was synthesized by Sigma-Genosys Ltd. Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich, all restriction enzymes were obtained from the TaKaRa Biotechnology Co. Ltd., and all cell culture products were purchased from Gibco BRL/Life Technologies, a division of Invitrogen. HBsAg ELISA Kits were purchased from the SINO-American Biotechnology Company. Purified HBsAg protein was generously provided by the Wuhan Institute of Biological Products.
-
HepG2 cells were grown in DMEM/High sup-plemented with 10% fetal bovine serum. HepG2.2.15 cells were grown in DMEM/High supplemented with 10% fetal bovine serum and 500μmol/L G418. All cell lines were grown in an incubator at 37℃ with an H2O-saturated 95% air-5% CO2 atmosphere. Cells were subcultured (Trypsin-EDTA) at ratios of not less than 1 to 3, and were grown in 35-mm plastic dishes for 1d prior to most experiments.
-
Purified HBsAg protein was separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and stained using a Silver Stain Plus kit (Bio-Rad Laboratories, Hercules, CA).
-
Repeated rounds of aptamer selection and amp-lification were performed as described previously [33]. In brief, the oligonucleotide template was synthesized as a single-stranded 115-mer with the sequence5'-TTAATACGACTCACTATAGTTGATTGCGTGTCAATCATGG-25N-GGTCATGTGTATGTTGGGGA TTAGGACCTGATTGAGTTCAGCCCACATAC-3', where the central 25N represents random oligonu-cleotides based on equal incorporation of A, G, C, and T at each position; the sequence of the T7 polymerase promoter was underlined. The dsDNA molecules were generated by PCR amplification using primer 1 (5'-TTAATACGACTCACTATAGTTGATTGCGTGTCAATC-3') and primer 2 (5'-GTATGTGGGCTGAA CTCAAT-3'). The initial RNA random library was prepared by T7 in vitro transcription at 37℃ for 4-16 h in a 50μL reaction mixture (0.5 to 1μg DNA, 5μL 10×transcription buffer, 400μg/μL PEG, 0.5mol/L MgCl2, 100U/mL IPP, 50 U RNasin, 2.5mmol/L CTP, 2.5mmol/L UTP, 2.5mmol/L ATP, 2.5mmol/L GTP, and 0.36μg/μL T7 RNA polymerase). Products were then treated with 10 units of DNase I for 30 min at 37℃ and stopped with EDTA to 20 mmol/L. Products were separated by 6 mol/L urea/8% PAGE, excised, purified, quantitated, and diluted into 1×binding buffer (20mmol/L Tris-Cl, pH8.0, 80 mmol/L Kac, 10% glycerol, 1mmol/L MgCl2, 0.2mmol/L EDTA, 10μmol/L. DTT).
Prior to each cycle, the RNA pool was passed through a HAWP filter (Millipore) pre-wetted with 1×binding buffer to remove potential filter-binding sequences. Pre-filtered RNA pool (final concentration to 300nmol/L) and HBsAg protein (final concentration to 30nmol/L) were mixed and the binding reaction was allowed to equilibrate for 40 min at 37℃ (1× binding buffer, 800μL final volume). The mixture was filtered through a HAWP filter pre-wetted with 1×binding buffer and rinsed with 5 mL 1×binding buffer. The filter membrane was then cut into small pieces and boiled in 200μL of elution buffer (7mol/L urea, 0.5mol/L ammonium acetate, 1mmol/L EDTA, 0.2% SDS) for 5 min. After a quick centrifugation, the supernatant was transferred to a fresh tube, and RNA was precipitated by adding 2.5 volume ethanol and 0.1 volume sodium acetate (pH 5.4). After being washed with 75% ethanol, RNA was redissolved in 20μL DEPC treated water.
After reverse transcription (RT)-PCR amplification with primer 1 and primer 2, the resulting DNA was transcribed with T7 RNA polymerase for 4-16h at 37℃ in a 50μL reaction mixture and the product was purified as described above. This purified RNA pool was used in the next round of selection.
-
After twelve rounds of selection, RT-PCR products were cloned into pMD19-T plasmid. The bank was transformed into E. coli DH5α. 24 clones were picked randomly, and plasmids were isolated from these clones, purified, and analyzed by sequencing.
-
RNA Structure Program version 4.6 (http://rna.urmc.rochester.edu/) was used to predict the secondary structures of aptamers from round 12. The most stable structures with the lowest free energies for each RNA oligo were compared.
-
RNAs were dephosphorylated by using alkaline phosphatase of calf intestine (New England Biolabs). After phenol extraction, dephosphorylated RNAs were 5'-termini labeled with [γ-32P] ATP by using T4 Polynucleotide Kinase (TaKaRa Biotechnology Co. Ltd.). All reactions were carried out according to the manufacturer's instructions for these enzymes.
-
10 nmol of 32P-labeled RNAs were allowed to equilibrate with an excess of protein HBsAg at 37℃ for 40 min. Samples were then filtered through a HAWP filter pre-wetted with 1×binding buffer and immediately rinsed with 5mL of the 1×binding buffer. The filter membranes were dried and counted by scintillation counting (Perkin-Elmer Micro Beta JET 1450LSC lumiscence counter).
-
Radiolabeled RNAs were mixed with protein HBsAg in 1×binding buffer and incubated at 37℃ for 40min. The mixtures were then loaded onto a 6% native PAGE gel and electrophoresed in 0.5×TBE buffer at 120V of constant voltage for 2h. The gel was directly analyzed using a Phosphorimager.
-
In vitro transcription was set up with or without 10mmol/L FITC-labeled UTP. The following was applied for a 50μL transcription reaction: 0.5 to 1 μg DNA template, 5μL 10×transcription buffer, 400μg/μL PEG, 0.5mol/L MgCl2, 100U/mL IPP, 50U RNasin, 2.5mmol/L 2F'-CTP, 2.5mmol/L 2F'-UTP, 2.5mmol/L ATP, 2.5 mmol/L GTP, and 0.36μg/μL T7 RNA polymerase, and treated water to obtain 50μL.
-
RNA pools of different rounds or aptamers from the final round pool were incubated with purified HBsAg protein in different molar ratio (10:1 or 100:1) at 37℃ for 30 min individually, the inhibition of the HBsAg reactivity by RNAs were then evaluated by enzyme-linked immunosorbent assay (ELISA). The detection of HBsAg by the commercial HBsAg ELISA kit was performed according to the manufacturer's instruction.
-
HepG2 and HepG2.2.15 cells were plated at a density of 1.0×105 cells/well or 1.5×105 cells/well respectively, for a 24-well plate one day prior to experiment. For the binding experiment, cells were fixed in 50% methanol for 15 min at 4℃ and dried at room temperature, stained with 50nmol/L FITC-labeled aptamer in PBS for 20 min at room temperature, and washed in PBS. All cells were imaged with Olympus IX70 Inverted Fluorescence Microscope and CoolSnap CF camera (Photometrics, Roper Scientific). Images were equally processed in Adobe PhotoShop (Adobe Systems Inc.) to increase brightness and contrast.
Materials
Cell culture
SDS-PAGE and Silver stain
In vitro selection
Cloning and sequencing
Predicting RNA secondary structure
32P Labeling of RNA
Nitrocellulose filter-binding assay
Gel mobility shift assay
In vitro transcriptions
HBsAg reactivity assay
Fluorescent staining
-
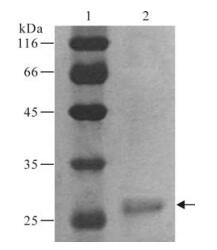
The HBsAg protein was found to be 95% pure according to the silver staining results which consisted of a single band of correct molecular weight (Fig. 1). The specificity of the protein was also confirmed by commercial HBsAg ELISA Kit.
-
Considering that aptamers would be applicable under some expected physiological conditions, they were selected to bind to HBsAg at 37℃ (pH 7.4) to approach in vivo conditions. A library of ~1.1×1015random-sequence RNA molecules was generated by in vitro transcription of a synthetic template and incubated for 40 min with purified HBsAg protein. Protein-nucleic acid complexes were isolated by filtration through a HAWP filter. The protein-bound RNA was then eluted and amplified by RT-PCR. The products of PCR amplification were then used as templates for in vitro transcription, generating a new pool of RNA enriched for HBsAg binding (Fig. 2A and 2B). Twelve rounds of selection and amplification were performed, and the ability of enriched RNAs to bind to HBsAg was monitored by nitrocellulose filter-binding assays (Fig. 2C) and gel mobility shift assay (Fig. 2D).
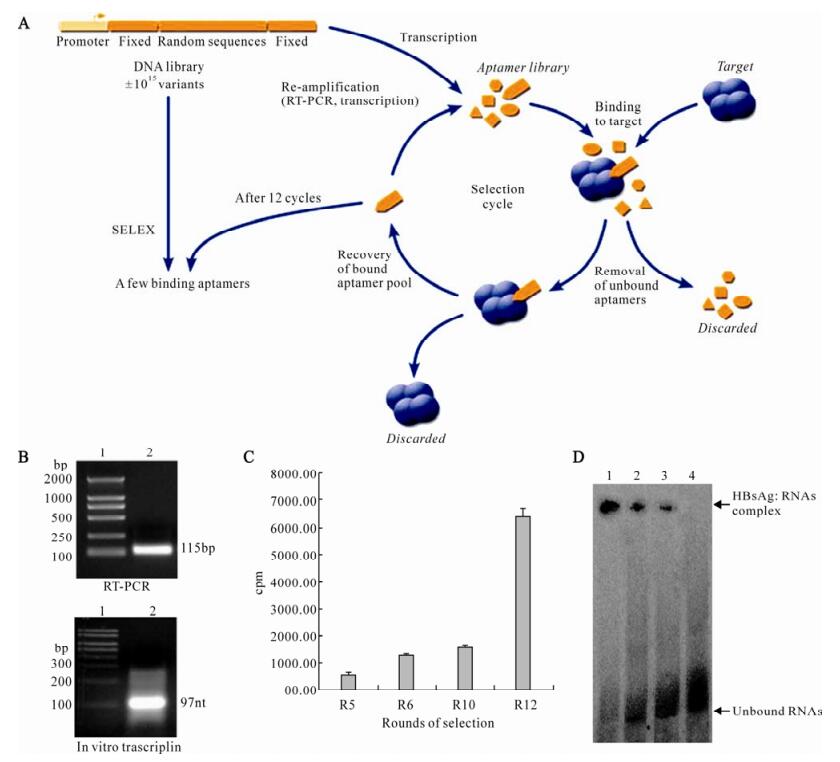
Figure 2. In vitro selection of HBsAg binding aptamers. A: Schematic of HBsAg in vitro selection. Repeated rounds of target binding, nitrocellulose filter selection, and amplification drove the evolution of the random sequence pool to bind to HBsAg protein. B: Enzymatic amplification of aptamers. RT-PCR: lane 1, DNA molecular weight marker; lane 2, PCR products in selex procedure. In vitro transcription: lane 1, RNA molecular weight marker. lane 2, Transcription products in selex procedure. C: Binding of RNA pools to HBsAg was increased with the ongoing selection process. Round 5, Round 6, Round 10, and Round 12 RNAs were 32P-labeled and incubated with HBsAg, binding of 32P-labeled RNAs were measured by Nitrocellulose filter-binding assay. D: Compelxes formation between RNA pools and HBsAg. 32P-labeled nucleo-protein complexes were separated from unbound RNAs by native gel electrophoresis. lane 1, Round 12 RNAs + HBsAg; lane 2, Round 10 RNAs + HBsAg; lane 3, Round 6 RNAs + HBsAg; lane 4, Round 0 RNAs + HBsAg.
-
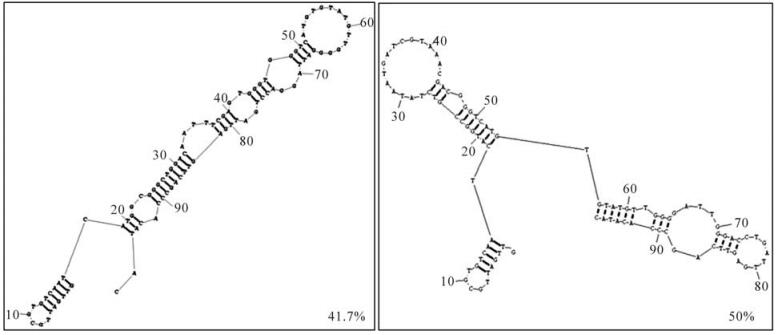
Round 12 RNAs were amplified by RT-PCR and cloned. Twenty four randomly picked plasmid clones were sequenced. The secondary structures of these aptamers were predicted with RNA Structure Program version 4.6. The most stable structures with the lowest free energies for each RNA oligo were compared. Over 90% of round 12 aptamers were represented by only two kinds of structures (Fig. 3).
-
In order to rapidly determine the binding affinity of the aptamers, HBsAg reactivity inhibition assays were performed. The original random sequence library had almost no effect on inhibition of HBsAg reactivity, whereas the inhibition ablilities of RNA pools increased with the ongoing selection process. The inhibition abilities of aptamers with typical structures from two different structure families were also assayed. One aptamer, termed HBs-A22, was found to possess maximum inhibition (Fig. 4).
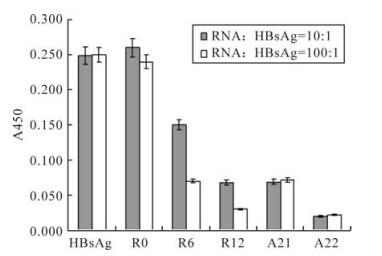
Figure 4. Aptamer inhibition of HBsAg reactivity. The original random sequence pool had almost no effect on inhibition of HBsAg reactivity, whereas the inhibition ablilities of RNA pools increased with the ongoing selection process, and the final round RNA pool can almost totally inhibit the HBsAg reactivity. Aptamer HBs-A22 was found having maximum inhibition than all the other aptamers.
-
Aptamer HBs-A22 was fluorescently labeled with FITC (HBs-A22-green) to evaluate if it could specifically bind to HBsAg-expressing liver cells. To increase nuclease stability of the aptamer in serum, 2'-fluro-pyrimidines were incorporated during aptamer trans-cription. Fluorescence microscopy showed that HBs-A22-green specifically bound to the HBsAg-positive hepatoma cell lines HepG2.2.15, while no binding was shown to the HBsAg-negative cell line HepG2, suggesting that the cellular binding of the aptamer depends on the presence of HBsAg (Fig. 5).
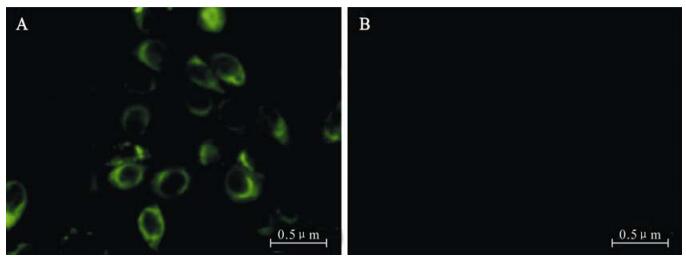
Figure 5. HepG2.2.15 cells staining by FITC-labeled aptamer HBs-A22-green specifically. Aptamer HBs-A22-green was incubated with HepG2.2.15 cells and HepG2 cells, the results were visualized by fluorescent microscopy. A: Aptamer H1-A25-green labels HBsAg-positive HepG2.2.15 cells. B: Aptamer HBs-A22-green shows no affinity for HBsAg-negative HepG2 cells.
Verification of HBsAg protein
HBsAg SELEX
Predicting Secondary Structures of Aptamers
Aptamer inhibition of HBsAg reactivity
Cell-specific binding
-
Aptamers are non-naturally occurring structured oligonucleotides that may bind to small molecules, peptides, and proteins. Typically, aptamers are generated by an in vitro selection process referred to as SELEX. High-affinity aptamers against over 150 different targets including ATP[31], cytokines[25, 26], growth factors[23, 28], proteases[1, 29], immunoglobulins[36], and cell adhesion molecules[14, 35] have been reported. Recently, aptamers have received validation as a therapeutic modality with approval of the anti-VEGF aptamer pegaptanib for the treatment of wet age-related macular degeneration (AMD)[3, 20]. Aptamers that bind with high affinity and specificity to proteins that reside on the cell surface have potential utility as therapeutic antagonists, such as being used for cell-type specific delivery of siRNAs[5, 15, 19]. In brief, the history of applications suggests that aptamers may soon be an attractive alternative to antibodies for both in vitro and in vivo applications. Despite the success of SELEX, there are no reported RNA aptamers that bind to HBV related antigens.
During HBV replication, high levels of HBsAg are expressed on the surface of HBV-infected hepatocytes. HBsAg is sometimes considered a "tumour-associated antigen". While membrane-associated HBsAg provides a target for direct immune-mediated cytolysis, it also provides a good target for the specific delivery of anti-hepatitis B drugs [16, 30, 32].
Therefore, we used purified HBsAg protein as a target to select apamers with high specificity and affinity. Filter binding assays and gel shift assays were performed to determine the specificity and affinity of the selected aptamers. Two kinds of aptamers with different structures were identified from an initial RNA aptamer library of ~1.1×1015 sequences. The high affinity of these aptamers is demonstrated by their ability to inhibit HBsAg reactivity. One unique HBsAg-binding aptamer, HBs-A22, was found to bind to protein HBsAg with higher affinity than the others. The secondary structure of aptamer HBs-A22 was similar to most other aptamers in the twelfth pool, as the sequences of randomized regions of HBs-A22 can form a stem-loop structure, which may participate in binding with protein HBsAg (Fig. 6). We also found that fluorescence-tagged aptamer HBs-A22 can bind to HBsAg-positive hepatoma cell lines HepG2.2.15 with high specificity.
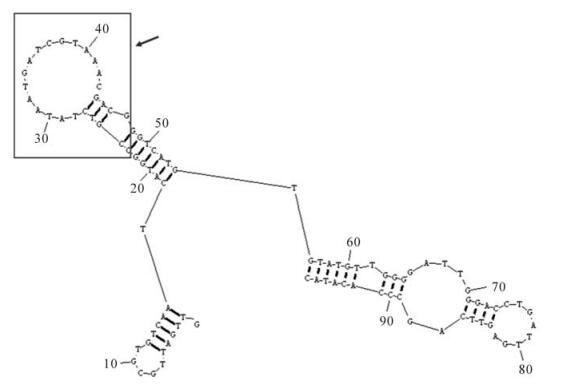
Figure 6. Predicting secondary structure of aptamer HBs-A22. Nucleotides 23-47 of aptamer HBs-A22 (box) represented the sequences selected from randomized region of RNA library which can form a stem-loop structure.
The work here describes the first application of SELEX to a HBV specific antigen and the first HBsAg aptamer. Additional modification of this high affinity aptamer toward even higher affinity and nuclease resistance should allow it to serve as a delivery vehicle for imaging, diagnostics, and therapeutic agents to the HBV related diseases.













 DownLoad:
DownLoad: