-
Severe acute respiratory syndrome coronavirus (SARS-CoV) is the etiological agent of SARS which emerged in Guangdong province, China during 2002 and 2003 [7, 17, 22, 23, 27, 36]. Closely related viruses were identified in masked palm civets (Paguma larvata), raccoon dogs (Neyctereutes procyonoides) in wild markets, suggesting that SARS-CoV is a zoonotic virus which was recently transmitted to human[9]. Subsequently, a group of genetically diverse CoVs, named SARS-like CoV (SL-CoV), was discovered in horseshoe bats (genus Rhinolophus), further demonstrated that bats may be natural reservoirs of SARS-CoV [18, 20].
Bat SL-CoV and human SARS-CoV share an identical genome organization and high sequence identities in gene products (92%-100%) with the main exception of the N-terminus of the spike glycoprotein (S protein) which shares only 64 % amino acid sequence identity, implying that the bat SL-CoV S protein has different properties from that of SARS-CoV [20]. The S protein of SARS-CoV is responsible for binding the cellular receptor, angiotensin-converting enzyme 2 (ACE2) [19], and mediates viral entry[6, 16, 28]. Additionally, the S protein induces protective humoral and cellular responses in the host[4, 5, 10, 11, 13-15, 21, 32, 34, 37]. For the bat SL-CoV S protein, by using the HIV-pseudotyped SARS-CoV and SL-CoV S protein, we have demons-trated that SL-CoV could not use ACE2 as its receptor. However, the replacement of the SARS-CoV receptor binding domain (RBD) to SL-CoV S protein could convert the SL-CoV pseudovirus to be susceptible to the human ACE2 expressing cells[26]. Furthermore, our previous study showed that viral-like particles containing the SL-CoV S protein have a stronger ability to stimulate dendritric cells (DCs) to cytokine induction than those containing the SARS-CoV S protein. And SL-CoV S DNA vaccine evoked a more vigorous antibody response and a stronger T cell response than the SARS-CoV S DNA in mice[1]. These results showed that the SL-CoV S protein has properties that are distinct from SARS-CoV S protein. Thus it is of great importance to elucidate its immuno-genicity.
In this study, we constructed a recombinant adenovirus expressing the full-length codon-optimized S gene of bat SL-CoV Rp3 isolate (rAd-Rp3-S), and investigated its ability to induce humoral and cellular immune responses in mice. The ELISA data showed that the rAd-Rp3-S immunized mice generated strong humoral immune responses against the HIV-pseudotyped SL-CoV S protein. However, the induced antibody had weaker cross-reaction with the HIV-pseudotyped SARS-CoV S protein; neither neutralized the pseudovirus. ELISPOT assay found that the secretion levels of IFN-γ and IL-6 in the rAd-Rp3-S immunized mice were higher than that of the negative control, demonstrating that the cellular immune responses were also elicited in the rAd-Rp3-S immunized mice. Our study demonstrated that the S protein from bat SL-CoV can elicit effective immune responses. It also suggested that rAd-Rp3-S can be a potential vaccine candidate against the group of SL-CoVs in the future.
HTML
-
The AdEasy adenoviral vector system (Stratagene), comprising the shuttle vector pAdTrack-CMV and the backbone vector pAdEasy-1, was used to construct a recombinant adenovirus carrying the full length codon-optimized S gene of bat SL-CoV Rp3 isolate (GenBank accession NO.DQ071615)[26], named rAd-Rp3-S. Briefly, the full-length codon-optimized S gene of bat SL-CoV Rp3 isolate was amplified from the plasmid pcDNA3.1-Rp3-S with primers 5'-GTAGTCGACACCATGGACGCCATGAAGAGGGG-3', and 5'-GTACTCGAGTCATCTTTTCTCAGCCAT CGC-3' (Sal I and Xho I sites are underlined) and subcloned to pAdTrack-CMV through Sal I and Xho I sites. Secondly, pAdTrack-CMV carrying the S gene was digested by Pme I, homo recombined with pAdEasy-1 to generate pAd-Rp3-S. Lastly, rAd-Rp3-S was obtained by transfecting PacI-linearized pAd-Rp3-S into 293 cells. In parallel, rAd-GFP, a recombinant adenoviral vector containing the GFP gene, was constructed according to the same procedure and used as vector control. Both of the two recom-binant adenoviruses were observed under florescent microscopy and titered with GFP marker. The virus was purified by CsCl-banding and observed under electronic microscopy (EM).
-
293 cells were maintained in DMEM supplemented with 100 IU/mL of penicillin and 100 μg/mL of streptomycin and 10% fetal calf serum. To detect S gene transcriptional level, total RNA of 293 cells infected by recombinant adenoviruses were extracted using TRIZOL® reagent (Invitrogen), then treated with DNase I (RNase free) at 37℃ for 30 mins. The first strand cDNAs were synthesized with MLV Reverse Transcriptase with random primers (Promega). Subsequent PCR was performed with primers specific for the codon-optimized S gene of Rp3, Rp3SF (5'-C GCGGATCCACCATGGCCCAGGAGGGCTGC-3') and Rp3SR (5'-CCGGAATTCGAGACATAGTGTA AGCCAC-3') to generate a 1956 bp fragment. The β-actin was amplified with primers AF (5-tggcg cttttgactcaggat-3) and AR (5'-agccctg gctgcctcaac-3') to produce a 450 bp fragment that was assayed as an internal control. RT-PCR products were analyzed by 0.8% agarose gel.
-
293 cells were harvested at 48h post infection and lysed in SDS buffer (50 mmol/L Tris.HCl, pH6.8, 100 mmol/L DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol). Lysate protein were fractionated on 4%-10% SDS-PAGE and transferred to polyviny-lidene difluoride (PVDF) by semi-dry protein transfer apparatus (Bio-Rad, Hercules, CA).The membrane was blocked for 2 h with 3% skimmed milk in TBS buffer (20 mmol/L Tris base, 137 mmol/L NaCl, pH 7.6). Following, the blocked membrane was probed with F26G8, mouse IgG mono-antibody against the SARS-CoV S protein that was kindly provided by Dr. Berry[3] and bound with goat anti-mouse AP-con-jugated antibody (Santa Cruz Biotechnologies, Santa Cruz, CA). The results were finally revealed by using chemical substrate NBT and BCIP coloration.
-
The six-week-old female BALB/c mice (n = 5 per group) were intraperitoneally immunized every 2 weeks with rAd-Rp3-S (1×107 pfu per dose) or with the negative controls (rAd-GFP or PBS). Mice were immunized three times for each condition before bleeding.
-
Sera from the immunized mice of different groups were collected on day 0, 14, 28 and 42 d. S protein specific antibodies (IgGs) were titered by ELISA. Firstly, HIV/BJ01-S and HIV/Rp3-S, pseudotyped with SARS-CoV (BJ01 isolate, AY278488) and SL-CoV (Rp3 isolate, DQ071615) S proteins, were produced as described previously[26]. The two pseudoviruses were used antigens and coated in separate 96-well microtiter plates. Then, the plates were incubated at 4℃ overnight and blocked with PBS (pH 7.4) containing 3% skimmed milk at 37℃ for 2 h. Mouse sera, diluted 500-fold, were added to the wells and incubated at 37℃ for 1 h. Wells were rinsed five times, 3min per time with PBST (PBS containing 0.05% Tween-20), followed by incubation with horseradish-peroxidase -conjugated goat-anti-mouse IgG (Sigma) (1:5 000) at 37℃ for 45 min and a subsequent 15 min incubation with the substrate TMB solution at room temperature in dark. Reactions were terminated with a stop solution, and optical densities (OD) were determined using a microplate reader set at 450 nm.
-
Pseudovirus-based neutralization assay was used to determine neutralization ability of immunized sera to HIV/BJ01-S. The neutralizing activity of heat-inactivated sera (56℃, 30min) was determined by mixing 10 ng of pseudovirues (in 30μL) with diluted antisera (in 30μL) at 37℃ for 1 h. Antisera-pseudovirus complexes were then mixed with 16 ng polybrene (in 40 μL medium) before they were added to human ACE2 expressing HeLa cells. The infected cells were washed with PBS and lysed (Cell Culture Lysis Reagent, Promega) at 48 h post infection. The neutralization activity of each antiserum was monitored by measurement of luciferase intensity as described previously [26]. Due to a lack of permissive cell lines for bat SARS-like CoV, the sera neutrali-zation assay to this virus was not conducted.
-
To test cellular immune responses in mice, ELISPOT assays were performed to detect the secretion levels of IFN-γ and IL-6 following the protocol of the manufacturer (U-CyTech, Netherlands; eBioscience, USA). Briefly, 96-well a ELISPOT plate was coated with 100μL anti-mouse IFN-γ or IL-6 capture antibody solution per well and left overnight at 4℃. The next day, wells were washed and blocked with complete RPMI-1640 at room temperature for 2 h. Splenocytes (2×105 per cell) from mice immunized for 56 days were added to microwells along with purified Rp3-S protein (10μg/mL) as the specific stimulating antigen. Cells were incubated at 37℃ and 5% CO2 for 36 h. Then wells were extensively washed with PBS containing 0.05% Tween 20 and subsequently incubated with biotin-conjugate anti-mouse IFN-γ or IL-6 detection antibody solution at room temperature for 2 h. After washing, wells were incubated with streptavidin-horseradish peroxidase antibody solution at room temperature for 45 min. Wells were washed again, and the freshly-prepared AEC substrate solution was added to wells. Color development was monitored for 10-60 min and stopped by washing with distilled water. After air-drying the plate at room temperature, wells were counted using an ELISPOT reader (Hitech Instruments).
Construction and purification of rAd-Rp3-S
Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Western blot analysis
Immunization of mice
Enzyme-linked immunosorbent assay (ELISA)
Sera neutralization assay
ELISPOT assay
-
The recombinant adenovirus, rAd-Rp3-S was observed by EM. The recombinant virus displayed a typical morphology characteristic of adenovirus (data not shown). The titers of rAd-Rp3-S and rAd-GFP, determined by GFP fluorencence, were up to 6.45×107PFU/mL and 5.53×107PFU/mL, respectively.
-
Green fluorescence was observed in rAd-Rp3-S infected cells at 24 h post infection. The RT-PCR result showed that an expected 1956bp fragment of bat SL-CoV S gene was obtained only in 293 cells infected by rAd-Rp3-S (Fig. 1A). An expected 450 bp fragment specific for β-actin was produced in 293 cells infected by rAd-Rp3-S and rAd-GFP (Fig. 1A). For western-blot, an expected 150-200kDa fragment that was in agreement with the size of SL-CoV S protein was observed in 293 cells infected by rAd-Rp3-S, but was not found in cells infected by rAd-GFP (Fig. 1B).
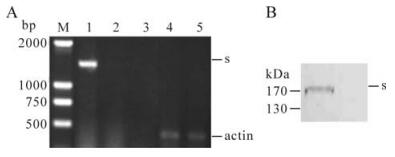
Figure 1. Expression of bat SL-CoV S protein in rAd-Rp3-S infected 293 cells. A: 293 cells were infected with rAd-Rp3-S and rAd-GFP respectively for 48 h and total RNAs were extracted for RT-PCR analysis. Lane 1 and 4, 293 cells infected with rAd -Rp3-S; Lane 2 and 5, 293 cells infected with rAd -GFP; Lane 3, the distilled water was as the blank control. B: 293 cells were infected with rAd-Rp3-S and rAd-GFP respectively for 48 h, the infected cells were collected and lysed for western blot analysis. Lane 1, 293 cells infected with rAd-Rp3-S; Lane 2, 293 cells infected with rAd -GFP. Arrow indicates the specific S protein.
-
The mouse sera collected on day 0, 14, 28, and 42 was used to detect specific antibody responses against HIV/BJ01-S and HIV/Rp3-S. ELISA results showed that the specific antibody against HIV/BJ01-S or HIV/ Rp3-S was detected in rAd-Rp3-S immunized mouse sera on the 14th day, the titer boosted dramatically after day 28 and was 10-fold higher than that in rAd-GFP immunized mice by day 42; no specific antibody responses were found at detectable levels in negative control groups, rAd-GFP or PBS immunized mice (Fig. 2). Further, the antibody titer against HIV/Rp3-S was much higher than that against HIV/BJ01-S in rAd-Rp3-S immunized mice. These results indicated that the specific antibody against SL-CoV S protein and the weaker cross with SARS-CoV S protein were generated in rAd-Rp3-S immunized mice.
Figure 2. ELISA detection of specific antibody against bat SL-CoV S protein in immunized mice. Mice were immunized with, rAd-Rp3-S, rAd-GFP or PBS. Sera of immunized mice from different groups were collected on day 0, 14, 28, 42. The titers of antibodies against pseudovirus HIV/BJ01-S (A) and HIV/Rp3-S (B) were measured by ELISA. Data shown are the mean±S.D. of three experiments from the different group.
Due to lack of permissive cell lines for SL-CoV, HIV/BJ01-S which has a similar infectivity to wild SARS-CoV[26], was used to detect the neutralization of antibodies induced by rAd-Rp3-S. The cells infected by mixtures of HIV/BJ01-S and mouse sera induced by rAd-Rp3-S, rAd-GFP and PBS did not afford any protection. However, high neutralizing activity was found in mouse sera induced by the full length SARS-CoV S protein (BJ01-S) (Fig. 3). The serum neutralization assay demonstrated that the antibody induced by bat SL-CoV S protein couldn't neutralize the HIV/BJ01-S and so the recombinant adenovirus rAd-Rp3-S may not be a cross protective candidate for SARS-CoV.
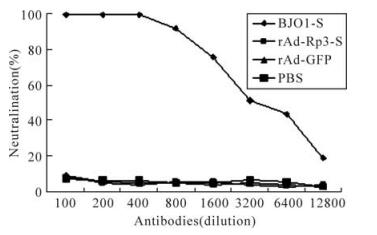
Figure 3. Neutralization test of antibody induced by rAd-Rp3-S. The antibody against BJ01-S, rAd-Rp3-S or rAd-GFP (dilution at 1: 100 to 1: 12 800) was mixed with HIV/BJ01-S and tested the infectivity to HEK293 cells expressing human ACE2. Percentage of neutralization activity was calculated for each antiserum as described in reference[26], and the average values were plotted.
-
The splenocytes of the immunized mice that were harvested on the eighth week were used to detect the secretion levels of IFN-γ and IL-6 by ELISPOT assay. The secretion numbers of IFN-γ were about 120 cells per 106 cells in rAd-Rp3-S immunized mouse splenocytes. However, the secretion numbers of IFN-γ were less than 10 cells per 106 cells in Ad-GFP or PBS immunized mouse (Fig. 4A). Similar to IFN-γ, the secretion numbers of IL-6 were up to about 160 cells per 106 cells in rAd-Rp3-S immunized mouse splenocytes, while only about 60 IL-6 secretion cells per 106 cells in Ad-GFP or PBS immunized mouse (Fig. 4B). These data demonstrated that IFN-γ, Th1 cytokine, and IL-6, Th2 cytokine, were both elicited in the rAd-Rp3-S immunized mouse and bat SL-CoV S protein can generate specific cellular immune responses in immunized mice.
Figure 4. ELISPOT detection of cytokine in immunized mice. The secretion levels of IFN-γ (A) and IL-6 (B) were detected in mouse splenocytes immunized by rAd-Rp3-S, rAd-GFP or PBS on day 56 post immunization. Data shown are the mean±S.D. of three experiments from the different group. SFC means spot forming cell.
Characterization of rAd-Rp3-S
Expression of S protein in rAd-Rp3-S infected 293 cells
Specific antibody responses in rAd-Rp3-S immunized mice
The cellular immune responses in rAd-Rp3-S immunized mice
-
The discovery of genetic diverse bat SL-CoVs in nature poses a potential threat to domestic animals and humans. However, little is known about this group of viruses, for example, the biological property, patho-genesis and immunogenicity. In this study, we successfully constructed a recombinant adenovirus containing the full length codon-optimized S gene of the bat SL-CoV. The recombinant virus, rAd-Rp3-S, elicited strong antibody response in mice against SL-CoV S protein, but a low cross reaction with SARS-CoV (BJ01 isolate) S protein and failed to neutralize the HIV/BJ01-S protein. These results indicate that the recombinant virus can induce a specific antibody response against bat SL-CoV S protein in a mice model and the bat SL-CoV S protein has different immune responses from that of SARS-CoV. These results are consistent with our previous results that bat sera naturally infected by SL-CoV could not neutralize the human SARS-CoV [20].
The RBD of SARS-CoV S protein contains neutralizing epitopes and can induce neutralizing antibodies[5, 10, 13, 14]. The minor mutations in the S protein, particularly the receptor binding domain (RBD), abolished viral entry into the susceptible cells and failed to induce cross-protection neutralizing antibodies[12, 35]. An important difference between the bat SL-CoV and SARS-CoV S proteins occurs in the RBD, the RBD of bat SL-CoV S protein has two deletions (5 and 12-13 aa, respectively). This important difference led to different usage of the cellular receptor between SARS-CoV and SL-CoV; the bat SL-CoV cannot use ACE2 as its receptor[26]. Our current study further demonstrated that the divergence between the SARS-CoV and SL-CoV S protein also may cause the antibody induced by SL-CoV S protein to have a weaker cross reaction with SARS-CoV S protein and such that it did not neutralize HIV/ BJ01-S.
Cellular immune responses are very important for host defense against the viral infection. IFN-γ is synthesized almost exclusively by activated natural killer (NK) and T cells in response to viral infection [24]. IL-6 plays a central role in both innate and acquired immune response. In our study, high secretion level of IFN-γ (120 cells per 106 cells) and IL-6 (160 cells per 106 cells) in rAd-Rp3-S immunized mice splenocytes, indicated that both Th1 and Th2 cells were elicited in rAd-Rp3-S immunized mice. Similar responses have been observed in SARS patients and animal models, for example, the level of IFN-γ in SARS patients at the early stages of disease onset correlated with resolution of the viral infection[29], and IL-6 was up-regulated as a consequence of being induced by SARS-CoV S protein in murine macrophages[31].
It is clear that the recently discovered bat SL-CoVs are not the immediate progenitors of SARS-CoV which caused the outbreaks in 2002/2003[26]. It is envisaged that the ongoing vaccine against the SARS-CoV may not protect the infection by bat SL-CoVs. Considering the wide distribution and genetic diversity of bat SL-CoVs in China[18, 20, 25, 33], co-infection of same bat species by different coronaviruses[18, 20, 30] and the capability of recombination of coronaviruses[2, 8], it is likely new coronaviruses in bats that can cross species to infect humans. Moreover, high density of bat habitats and increasing contacts between bats and human will further increase the virus transmission opportunities from bats to human. So, it is highly important to be prepared for prevention and control of the emerging disease resulting from this group of viruses. The recombinant adenovirus constructed in this study provides a potential vaccine candidate against the infection by diverse bat SL-CoVs.













 DownLoad:
DownLoad: