-
Herpes simplex virus type Ⅰ (HSV-1) is a double-stranded DNA virus with a complicated structure and a specific role in the cascade effect in gene transcription regulation. HSV-1 shows characteristics at different levels in its interaction with cells [2, 11], such as the influence of HSV-1 infection-related protein on viral replication function—the working mechanism of which has gradually become one of the key research fields in molecular biology [6, 10]. In our previous studies, we conducted more detailed research on related proteins induced by HSV-1 infection in human fibroblasts using mRNA differential display technology and protein 2D differential technology [7, 8], and obtained a number of HSV-1 infection-related proteins [9, 13]. Once such protein was HTRP (human transcription regulator protein) which is associated with transcriptional regulation and interacts with part of the transcriptional co-repressor mSin3A complex SAP30 (mSin3A Association Protein) to promote HSV-1 infection which leads to cell death [5]. This finding suggested that HTRP may have a direct or indirect impact on HSV-1 transcriptional regulation as an HSV-1 infection-related protein, possibly through some specific intracellular biological pathway. Our research could potentially provide a direct basis for helping to understand the biological significance of the interaction between HSV-1 and host cells, reflected in the regulation and transcription of viral genes through an intracellular protein modification system. On this basis, further experiments were designed to analyze the transcriptional regulation mechanism of different HSV-1 genes by HTRP. The results showed that HTRP's influence on the transcription regulation process of HSV-1 was simulated by promoting deacetylation enzyme HDAC activity, which regulates viral gene transcription through HSV-1 gene chromatin reunion/deconstruction. The results provide some insight into the biological characteristics of using the intracellular chromatin reunion/deconstruction mechanism to achieve different pathological results in the process of HSV-1 infection. In addition, a relevant research basis would be provided for understanding the interaction between HSV-1 and its host cells from the view of cellular molecule modifying enzymes.
HTML
-
The cryopreserved 293 cell line was recovered under conditions of 37℃, 5% CO2 and 95% relative humidity, and was cultured in complete culture medium DMEM (plus 10% newborn calf serum, 2 mol/L glutamine, 100 mg / L penicillin and 100 mg/L streptomycin). Plasmids pCAT-TK, pCAT-gC and pcDNA3-HTRP were preserved by our laboratory. Plasmids pcDNA3, pGL3-Basic, pRL-CMV were obtained from Promega.
-
The total RNA was extracted from 293 cells using Trizol reagents. Using reverse transcriptase M-MLV, the first strand cDNA was synthesized and used as template with primers P1 and P2 for PCR amplification under the conditions of 98℃ 10 s, 60℃ 5 s, 72℃ 1 min (30 cycles), then 72℃ 10 min, 4℃ incubation, then human SAP30 gene was obtained. SAP30 gene primers sequences were P1: 5'-TATAA GCTTATGAACGGCTTCACGCC-3'; P2: 5'-AGCGA ATTCCTA GTGAACACCACTAT-3'.
The recombinant plasmid pcDNA3-SAP30 was constructed by inserting a fragment into the pcDNA3 vector through a double enzyme digest. The recombinant plasmids pGL3-TK and pGL3-gC were constructed by cutting TK and gC fragments from pCAT-TK and pCAT-gC plasmids respectively, and ingesting the pGL3 vector with the single restriction enzyme Bgl Ⅱ.
-
The cells were inoculated into 10 cm culture flasks and were not used until they grew to 80%-90%. Lipofectamine 2000 was used to transfect with the corresponding plasmid in accordance with the manufacturer's instructions. The cells were harvested for follow-up experiments after being cultured at 37℃ in a 5% CO2 incubator for 24-48 h.
-
The plasmids pcDNA3-HTRP and pcDNA3 were transfected into 293 cells, which were scraped after being washed three times with PBS for 40 h later. The split cells were put in an ice bath with RIPA lysis buffer for 30 min and protein supernatant was collected by centrifugation. The same amount of protein supernatant was taken for SDS-PAGE electrophoresis, transferred to PVDF membrane and closed in 5% skim milk TBST with HTRP anti-serum (prepared by our laboratory) or SAP30 antibody as the first antibody, a sheep anti-mouse IgG-HRP as the secondary antibody and ECL as colour light-emitting agents.
-
293 cells grown in.6-well plates were transfected with the corresponding plasmid and 24h later, were inoculated with HSV-1 virus at a concentration of MOI = 1. The cells were harvested after infection at 4 h, 6 h and 8 h. Then quantitative real-time PCR reaction was used to detect gene mRNA level using TK, gC or β-actin as primers. Primers were as follows: TK-F: 5'-TGACTT ACTGGCGGGTGTTG-3', TK-R: 5'-GGTCGAGGCGGTGTTGTG-3'; gC-F: 5'-GATGC CGGTTTTGGAATTC-3', gC-R: 5'-CCCATGGAGT AACGCCATATCT-3'; β-actin-F: 5'-ACACCCCAG CCATGTACGTT-3', β-actin-R: 5'-TCACCGGAGTC CATCACGAT-3'. Reaction conditions were 95℃ 30 s, 95 ℃ 5 s, 56 ℃ 10 s, 72 ℃ 40 s, for 40 cycles. The final result was obtained by averaging three independent experiments.
-
293 cells were inoculated into 24-well plates and when the integration degree reached 70-80%, the mixed plasmids of expressed pcDNA3-HTRP, pcDNA3-SAP30, pcDNA3, pcDNA3-HTRP and pcDNA3-SAP30 were co-transfected with pGL3-TK or pGL3-gC and the control group was transfected into pGL3-Basic. All cells were co-transfected with pRL-CMV to provide the internal control of the transcriptional activity. Cells were harvested 40 h after transfection. TSA (10ng/mL) or DMSO was added after the transfection. The detection of reporting gene activity was performed in accordance with the dual fluorescein detection kit instructions. All experimental groups were compared to the control group to get the relative luciferase activity (RLA). Each experiment was repeated three times and the RLA mean was calculated for statistical analysis.
-
Plasmid pcDNA3, pcDNA3-HTRP and pCAT-TK or reporter plasmid pCAT-gC were co-transfected into 293 cells respectively and CHIP analysis was conducted 40 h later. The cells were cross-linked with formaldehyde, washed with PBS, scraped and treated with ultrasound. Immunoprecipitation reaction was carried out with moderate anti-acetylated histone H3K9 or H3K14 antibody and the complex was precipitated with agarose protein A/salmon sperm DNA. The washed, eluted, anti-cross-linked and purified DNA was detected by Real-Time PCR. Primers were TK promoter-F: 5'-TGCGAAGTGGACCTGGGAC-3', TK promoter-R: 5'-CCGTTGCTCGCG TTTGCT-3'; gC promoter-F: 5'-CAATAAAAGGCATTAGTCCC G-3', gC promoter-R: 5'-ACCAAGCGCCCTCGATA GTG-3'; β-actin promoter-F: 5'-TGCTGCACTGTGC GGCG-3', β-actin promoter-R: 5'-GTCTCGGCGGTG GTGGC-3'; the negative control primer-F was 5'-GCC ATTCGCCATTCAGG-3'; the negative control primer -R was 5'-ATTTTATTGCGG CCGCTC-3'. Real-Time PCR reaction conditions were the same as described above.
-
The experimental data were expressed with mean ± standard deviation. A T-test analysis was performed in both the experimental and the control groups, with a result of P < 0.05 consider to an indication that the data are statistically significant.
Cells and plasmids
Construction of recombinant plasmid
Cell transfection
Western blot detection
Detection of TK, gC gene mRNA level by quantitative real-time PCR
Analysis of dual-luciferase activity
Chromatin immunoprecipitation (CHIP)
Statistical analysis
-
Based on the analysis of the cellular function of HTRP protein in our previous study [5], the transfection method was further used to prepare the cell environment for up-regulation of HTRP expression in human 293 cells. In addition, part of the biological process of HSV-1 infection and replication was analyzed. First, we used Western blot methods to identify HTRP protein expression background in human 293 cells after being transferred with pcDNA3-HTRP. We observed the impact on the proliferation dynamic process of HSV-1 infection by the up-regulated HTRP. The results show that HTRP expression could be obtained in human 293 cells using pcDNA3 as the carrier (Fig. 1A). After inoculation of HSV-1 (MOI = 1) into the cells, we analyzed the transcription of viral TK and gC genes and found that HTRP had an obvious inhibitory effect on the transcription of these two viral genes (Fig. 1B). The inhibitory effect showed an increasing trend with the extension of infection time (Fig. 1B).
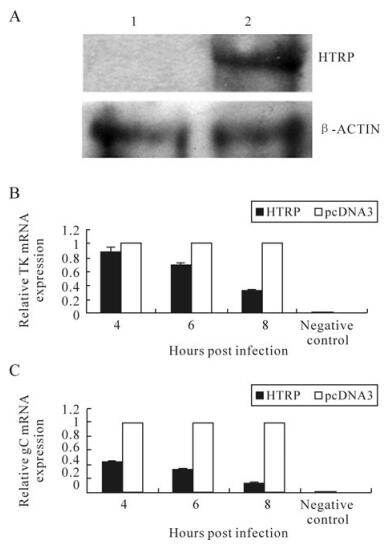
Figure 1. Effect on HSV-1 TK and gC gene transcription by HTRP expression up-regulation. A: The detection of HTRP expression in 293 cells by Western blotting: 1. Transfected with pcDNA3; 2. Transfected with pcDNA3-HTRP. mRNA expression changes were detected using Real-Time Quantitative RT-PCR and the result was calculated by the formula: lg2-△△Ct. B: expression level of TK mRNA. C: expression level of gC mRNA.
-
The results in our earlier experiments proved that HTRP can interact with the SAP30 molecule of the intracellular transcriptional repression complex mSin-3A and thereby promote the trend of cell death [5]. To further explore whether the interaction between HTRP and SAP30 would produce a synergic effect on HSV-1 transcription, 293 cells were prepared for the co-transfection and expression of HTRP and SAP30 in our experiments. After determining the up-regulated expression of these two molecules in cells (Fig. 2A), the transcriptional efficiency of the HSV-1 TK gene in the infected cells was observed using real-time quantitative PCR. The same inhibitory effect on TK gene transcription was demonstrated by up-regulating HTRP or SAP30 expression in cells (Fig. 2A). However, the inhibitory effect on TK gene transcription was more prominent in the cells co-transfected with HTRP and SAP30 (Fig. 2B). This result suggests that HTRP and SAP30 have an inhibitory synergic effect on the transcription of the TK gene. Similar results were obtained for the transcription of the gC gene (Fig. 2C).
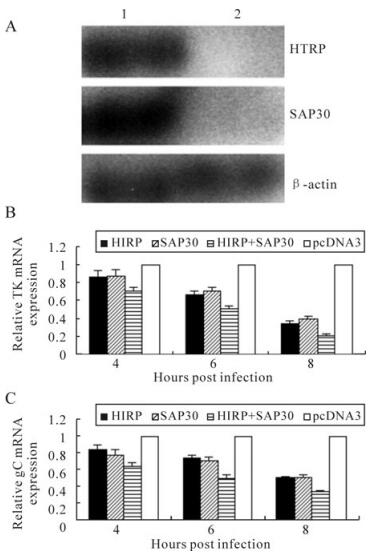
Figure 2. Synergic inhibitory effect of HTRP and SAP30 on TK gene and gC gene transcription. A: The detection of HTRP and SAP30 expression in 293 cells by Western blotting: 1. transfected with pcDNA3-HTRP and pcDNA3-SAP30; 2. transfected with pcDNA3; mRNA expression changes were detected using Real-Time Quantitative RT-PCR and the result was calculated by the formula: lg2-△△Ct. B: expression level of TK mRNA. C: expression level of gC mRNA. Indicates a significant difference (p < 0.05) as compared to controls.
-
Research on the transcriptional regulation of SAP30 and its mSin3A complex in the field of molecular biology indicates that co-transcriptional repression is usually accomplished through aggregating HDAC by Sin3A, leading to the deacetylation of histones H3 and H4 [4, 16]. SAP30 is an important structural component in this process, playing a specific role in raising and connecting HDACs to the complete Sin3A complex [12]. Therefore, in this sense, the synergistic effect of HTRP and SAP30 on the transcriptional repression of HSV-1 TK and gC gene is obviously related to the effect of HDAC enzyme activity. In our experiments, we further set the condition of the presence of HDACs inhibitor TSA to detect the transcriptional effect of HTRP and SAP30 on the HSV-1 TK and gC genes. The results showed that the presence of TSA could obviously relieve the transcriptional repression effect caused by HTRP, SAP30 and their synergy on the TK and gC genes (Fig. 3), suggesting that the transcriptional repression effect of HTRP and SAP30 alone or their synergy could be associated with the state of HDAC's enzyme activity.
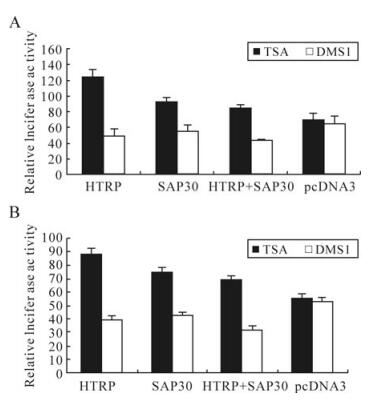
Figure 3. The effect of TSA on the inhibition of transcriptional activity of TK and gC promoter by HTRP and SAP30. The expressive plasmid were transfected into 293 cells respectively. The relative luciferase activity of each experimental group was obtained using the fluorescence activity of pGL3-Basic as a control. A: pGL3-TK relative fluorescence activity. B: pGL3-gC relative fluorescence activity.
-
The experimental results mentioned above demonstrate that HTRP's repression effect on the transcription of TK and gC genes is related to the enzyme activity of HDACs. Some experiments indicated that in the process of infection, the HSV-1 gene can be integrated with cellular histones to produce a chromatin phenomenon, and that the viral transcriptional activity can be regulated through the process of reunion and deconstruction of chromatin [3]. This means that HTRP may influence the transcriptional efficiency of the TK and gC genes through histone deacetylation. ChIP experiments were designed based on this inference and used specific primers of TK and gC gene sequences to analyze the deacetylation features of chromatin histone H3, combined with these two genes in HTRP-up-regulated cells. Our experimental results demonstrate that in the case of HTRP expression up-regulation, the formed chromatin H3 histone lysine 9 and 14 in TK and gC gene sequences showed significant deacetylation compared to the control (Fig. 4). The results show that HTRP promotes the role of HDAC's deacetylation, which is clearly educed by the combination with SAP30. An obvious deacetylation status appears in lysine 9 and 14 of histone H3 as a result.
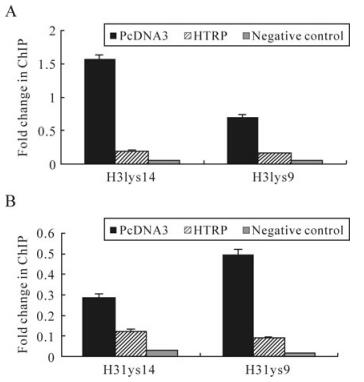
Figure 4. Impact of HTRP on the histone acetylation state of TK and gC gene promoters. Using the pcDNA3 transcription group as the control, Real-Time PCR was performed. The copy number ratio of antibody precipitated DNA to Input DNA was normalized to the copy number ratio of antibody precipitated β-actin DNA to Input β-actin DNA. The sequence that did not bind to the aimed protein in the target sequence upstream was used as the negative control and the acetylation modification relative value of each experimental group was obtained. A: TK promoter ChIP analysis. B: gC promoter ChIP analysis.
The up-regulation of HTRP expression can inhibit the transcription of TK and gC gene
Synergic inhibitory effect of HTRP and SAP30 on TK and gC gene transcription
Synergic inhibitory effect of HTRP and SAP30 on the transcription is related to HDAC
HTRP can promote the deacetylation of histone H3 at lysines 14 and 9
-
Complicated interactions exist between HSV-1 and its host cells; therefore, the pathological effects of different cell infections are diverse. This depends largely on the special transcriptional regulation mechanism of the viral gene and the interaction between viral components and cells during the process of infection and replication. In our previous work, we found that the immediate-early gene product HTRP protein induced by HSV1 infection in human fibroblasts could be combined with SAP30, one of the components of co-repressor complex mSin3A [9]. This is similar to another phenomenon: the RPMS molecule of EB virus can be combined with relative transcriptional repressor CIR of CBF1 to regulate the transcription of viral or cellular genes [15]. This mechanism can also be observed in the HCV virus [14]. These findings suggested that HTRP may also influence the transcription and regulation process of HSV-1 gene through a similar mechanism. It can be observed in our work that in the case of the expression up-regulation, HTRP can suppress the transcription of viral TK and gC gene at different infection phases. Furthermore, inhibition is more significant as the time is extended (Fig. 1). This clearly suggests that HTRP has a transcriptional repression function and can suppress the replication and proliferation of HSV-1 virus accordingly. We have confirmed in previous work that HTRP may be combined and interacted with SAP30. Under a cellular environment which contains simultaneous up-regulated expression of HTRP and SAP30, we further demonstrated that both showed a synergistic inhibition effect on the transcription of the TK and gC genes (Fig. 2). Cellular and molecular data have shown that as one of the major structural components of the co-repressor complex for the transcription and regulation in cells, SAP30 plays a role in mediating HDACs to be bound to the complex. The data have also shown that these can achieve transcriptional repression by chromatin histone deacetylation [12]. Therefore, the combination of HTRP and SAP30 and the synergic effect on the transcriptional inhibition of viral genes TK and gC suggests that HTRP, an up-regulated cellular protein induced by HSV-1 infection, may work through the HSV-1 gene chromatin reunion/ deconstruction. This may be related to the acetylation/ deacetylation mechanism to influence viral proliferation and replication processes. This inference was further confirmed by our TSA experiments using HDACs as an inhibitor (Fig. 3). Meanwhile, ChIP experiments also showed that the combination of SAP30 and HTRP caused changes in the activity of HDACs. Ultimately, the specific mechanism leading to the inhibition of viral gene transcription is that HDACs increases the degree of deacetylation of lysines 9 and 14 in histone H3 (Fig. 4). Previous experiments have shown that when HSV-1 enters into the cells, the histones of chromatin structures formed by combining with its gene include the H3 molecule [1]. Based on these data and according to the working molecular mechanism of HTRP up-regulated by HSV-1 infection in fibroblast cells, we propose the following model: As a transcriptional regulation molecule, HTRP is reactively up-regulated by the HSV-1 infection in cells and may function by combining with SAP30 to increase the enzyme activity of HDACs. These HDACs would function via the co-repressor complex Sin3A, up-regulating the deacetylation level of lysine 9 and 14 in histone H3 in regions binding to the HSV-1 gene promoter. As a result, the transcriptional process of HSV-1 related genes would be inhibited to some extent and the viral replication and proliferation would be affected. The establishment of this model along with experimental data will help us further understand the interaction between HSV-1 and host cells through a cellular protein modifying enzyme system to influence the transcriptional regulation of viral genes, which causes a variety of pathological results by HSV-1 infection in cells.













 DownLoad:
DownLoad: