-
Banana streak virus (BSV), a member of genus Badnavirus, is a worldwide pathogen which can cause streaked leaves on cultivated bananas (Musa spp.) and plantain plants [1]. The occurrences of BSV have been confirmed in banana production areas of Asia, Africa, Australia, Americas and Europe. BSV infection in Musa spp. causes reduction of fruit yield and quality. BSV can be naturally transmitted by the citrus mealybug (Planococcus citri Risso) or possibly other species of mealybug [11, 13]. However, inadvertent use of infected planting material for vegetative propagation which acts as a main avenue of propagating banana [3], have given rise to greater risk of dissemination of BSV than that caused by mealybug [2]. Therefore, the risk of BSV infection in bananas has become a hindrance to the international exchange of Musa germplasm. Moreover, some BSV species' genomes may integrate into the M. balbisiana host (B genome) [5, 9, 16]. In addition, tissue culture presents further risk for episomal expression of integrants and the induction of BSV infections; and other adverse environmental factors may also be involved in this virogenesis process [16]. In view of the risks associated with latent infections by BSV, there is a great need to detect BSV to achieve virus-free banana plantlets. However, the large-scale genetic divergency among BSV isolates can cause problems for PCR based detection of BSV.
To investigate the genetic characteristic of BSV infected banana plants in Yunnan province in China, we sequenced the entire genome of an isolate we named BSAcYNV from a plantation in Yunnan, and performed sequence and phylogenetic analysis to determine its relationship amongst other strains.
HTML
-
Naturally infected-banana leaves (Cavendish Musa AAA group) were sampled from Honghe, Yunnan province in China. Samples were frozen at -70℃ and purification of BSV was carried out using a modified method for BSV isolation as described previously [1]. BSV infected leaves eliminated midribs were cut into pieces and ground into powder in liquid nitrogen and blended in 400 mL of 0.2 mol/L phosphate buffer (pH6.0) per 100g banana leaves[containing 0.5% (w/v) sodium sulphite, 2% (w/v) PVP-40, and 10 mmol/L EDTA•Na2]. The extract were filtered through three layers of gauze, the filtrate was twice centrifuged for 20 min at 12, 000 r/min (JA 25 rotor) at 4℃ to remove plant debris. The NaCl and PEG were added into the upper supernatant to 1.5% (w/v) and 7.0% (w/v) of final concentrations respectively, and the mixture laid statically at 4℃ overnight. Then the mixture was centrifuged at 12, 000 r/min for 20 min. The precipitate was suspended in 10 mmol/L sodium phosphate buffer pH7.2 containing 0.9% (w/v) NaCl. Triton X-100 was added into the suspension to 0.1% (v/v) and stirred at 1h at 4℃, followed by centrifugation at 12, 000 r/min for 20 min. The supernatant was subjected to centrifugation under a 20% sucrose cushion at 45, 000 rpm for 2 h in a Beckman Type 55.2 rotor. The virus pellet was resuspended in the 10 mmol/L phosphate buffer pH7.2, and centrifuged at 12, 000 r/min for 10 min at 4℃. The virus preparation was used for further analysis.
-
Before BSV DNA extraction, the virus suspension was incubated with DNaseI at 37℃ for 30 min. The DNaseI was inactivated at 75℃ for 10min. Then the mixture was digested at 37℃ for 30 min with proteinase K in the presence of 1% (W/V) sodium dodecyl sulfate (SDS) [8]. The mixture was extracted with equilibrated phenol twice; nucleic acid from the aqueous phase was precipitated by 2.5 fold volume of absolute ethanol. The precipitate was collected by centrifugation, rinsed with 70% ethanol, dried under vacuum and resuspended in 20 μL TE buffer. The primers were designated based on the conserved sequences of the reported isolates, with forward primer pBSV1:5′-TGATATTC/TTTTC/TTG/TGAAAATCC-3′, and reverse primer pBSV2:5′-GAA/GTAGATGTCTGA G/TACA/GTC-3′ to generate a 673 bp fragment by PCR amplification. Primers for long-distance PCR amplification of the entire viral DNAs were designed using the PCR product sequence information. The back-to-back primers were contiguous, forward primer pCSeq1: 5′-CAAGTCACATGTGAGGATAG-3′ (sense); reverse primer-pCSeq2:5′-TGAGGCTTTCCTTTGTA CG-3′ (anti-sense). PCR was carried out in a volume of 50 μL containing 0.5 μL viral DNA, 0.4 μmol/L of each primer, 0.2 mmol/L dNTPs, 5 μL 10× LA Taq buffer, and 1.25 U LA Taq polymerase (TaKaRa). The amplification cycle conditions were 94℃ 3min, [94℃ 30s, 50℃ 30, 68℃ 7min30s] ×30 cycles, 72℃ 10 min. The PCR fragments were extracted using a QIA quick gel extraction kit. Then the purified PCR fragments were inserted in pMD18T (TaKaRa) and transformed into competent cells of E.coli DH5α.The recombination plasmid DNAs were extracted by the alkaline lysis method. Viral DNA inserts were sequenced from both ends with M13 forward and reverse universal primers and other following synthesized primers according to the last determined sequences by the ABI377 automated DNA sequencer (Applied Biosystems).
-
Multiple sequences were aligned by Clustal W (version 1.83) and the phylogenetic tree was constructed with MrBayes 3.2. The GenBank accession numbers of the sequences reported in this paper are as follows: BSIOV (from cv, Obino I' Ewai, Nigeria): ABI31268, BSMyV (from cv. Mysore, Australia): AAW80648, BSGfV (from cv. Goldfinger, Australia): AAR86692, BSAcVNV(from Acuminata, Vietnam): AAU95075, BSAcYNV (from cv. Cavendish, China): AAY99427; Commelina yellow mottle virus (ComYMV:CAA37110), Kalanchoe top-spotting virus (KTSV: AA021220), Sugarcane bacilliform virus (SCBV: AAA47454).
Sources of Virus and Virus purification
Cloning and DNA sequencing
Sequence Analysis
-
The Banana streak virus investigated in this study was from M. Acuminata and isolated in Honghe, Yunnan province, China. The virus purification gave a moderate yield of bacilliform particles of size 23±2 nm×110±10 nm (Fig. 1A), which was used to raise antibody against BSV (data not shown).
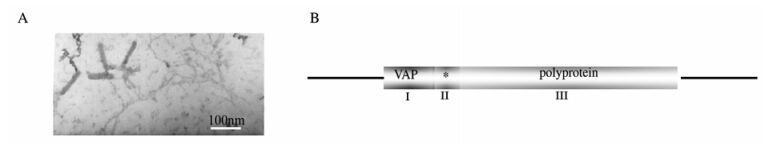
Figure 1. A: BSV particles visualized by transmission electron microscopy image shows bacilliform particles of size 23±2 nm×110± 10 nm. B: Genome structure of Badnaviruses. VAP:virion-associated protein, polyprotein conting MP (movement protein), CP (coat protein), PR (aspartic protease), RT (reverse transcriptase) and RH (RNase H) shown in.
The complete genome of BSAcYNV (Banana Streak Acuminata Yunnan Virus) consists of 7722 base pairs (bp). A linearized Badnavirus full-length genomic structure is shown in the Fig. 1B. Consistent with other Badnaviruses and Caulimoviruses, the numbering of the genome sequence begins at the putative 5'-terminal of the tRNAiMet-binding site. The genome of badnaviruses usually contains a single molecule of dsDNA of about 7.2-7.8 kb long that forms an open circle interrupted by site-specific discontinuities that could contain an intergenic poly (A) region [8, 9]. The virus genome usually contains 3 ORFs, whose functions remain to be elucidiated. The largest ORF 3 polyprotein product consists of the movement protein (MP), the virus coat protein (CP), the aspartic protease(PR), the reverse transcriptase(RT) and the ribonuclease H (RH) domains. The CP domain of badnaviral ORF 3 displays at the C-terminus a large region rich in zinc finger (CCHC) motifs [8, 9]. Additionally, some badnaviruses show an additional dUTPase domain upstream or downstream to the CP domain within ORF 3 similar to that of several Retroviridae retroviruses [16]. The function of the ORF 2 products remain elusive but the ORF 1-encoded protein is a virion-associated protein. The intergenic sequence is 1094 bp in the BSAcYNV genome (at positions 7538 to 7550) and was analyzed in silico to reveal a potential TATA box (sequence CTCTATAAGA GGA). The sequence TCACGCACGATGAC (7347-7363 bp) located upstream of the TATA-box is similar to the as-1 element in the 35S promoter of Cauliflower mosaic virus (CaMV). The CCA(N)nTGG motif and a repeated AGAAG motif (at positions 7244 to 7255) are also present in BSAcYNV promoter, which may impose vascular tissue-specific promoter activity, as observed in RTBV (Rice tungro bacilliform virus) and BSCvV (Banana Streak Cv. Virus) (from Cv. Williams, Australia) promoters [20]. A putative trans cription start site and a polyadenylation signal were predicted at position 7569 and at position 7625 respectively (Fig. 2). Hence, a terminally redundant RNA transcribed from the genomic DNA was revealed at the transcriptional level. This large RNA is greater than one genome in length, termed as the pregenomic RNA (pgRNA). The pgRNA begins with a long leader with a short ORF (sORF) preceding the first long viral ORF which serves as a polycistronic mRNA contain ning three large ORFs.
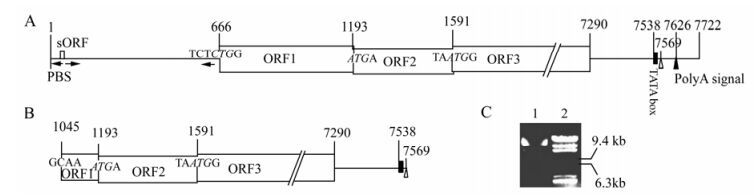
Figure 2. Comparison of the structure of complete and defective genomic DNA in BSAcYNV. The intergenic region is depicted as a line, open reading frames (ORFs) as boxes and numbers are the positions in BSAcYNV genome; the start codons of large ORFs are in italics; arrowhead adjacent to a vertical line shows the primer-binding site (PBS) for reverse transcription; a small black rectangle denotes the putative TATA box; white triangle indicates a transcriptional start site; black triangle indicates a putative polyA signal; arrows under intergenic region denote complementary sequences that form the base of the large stem-loop structure. A: Primary structure in + strand of complete genomic DNA. B: Primary structure in + strand of defective genomic DNA. This defective genome lacks the whole RNA leader region and the major ORF1 coding region. C: The exacted DNAs from BSV preparation represent different size bands in agarose gel. 1. BSV genomic DNAs; 2. λDNA/ Hind Ⅲ Marker.
In contrast to the complete genome, the defective genomic DNA sequence lacks the intergenic region and the majority of the coding region for ORF1. However, it still contains a TATA-box and transcriptional start site, so it can be putatively transcribed into RNA. The transcriptional start site (at position 7569) directly links up with the 3'-terminal coding sequence (at position 1045) of ORF1. Thereby, the truncated genomic DNA is composed of 6525 bp and devoid of 1197 bp of sequence. This may be consistent with several different morphological sizes of extracted BSV genomic DNA identified on agarose gel electrophoresis (Fig. 2).
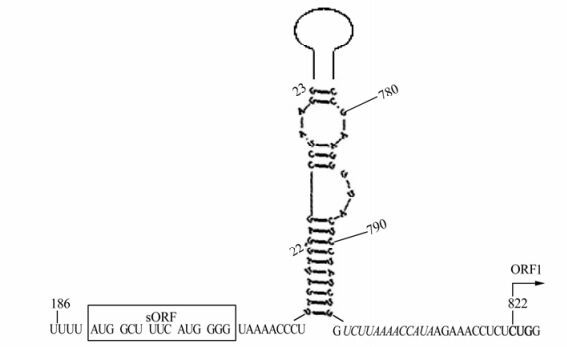
The folding of the BSAcYNV pgRNA leader resulted in an extended stem-loop structure formed just in front of ORF1, and only an in-frame CUG triplet (non-AUG start codon) in a favorable context was found in the downstream of AU rich sequence (Fig. 3), which could be used as a shunt acceptor. Also, ORF1 overlaps ORF2 using the motif AUGA, with AUG acting as the start codon of ORF2 and TGA acting as the stop codon of ORF1; ORF2 overlaps ORF3 via the motif UAAUG, with AUG acting as the start codon of ORF3 and UAA acting as the stop codon of ORF2. Along with the full-length mRNA, a fraction of shunting ribosomes could receive the AUG codon of ORF3 by leaky scanning. So we deduce that the combinational model of shunting and leaky scanning for pgRNA translation in BSAcYNV is similar to that in other Badnaviruses.
-
The positive strand contains three large ORFs, named ORF 1, ORF 2 and ORF 3. The ORF 1 of potentially encodes a protein of 176 amino acids. Note that the ORF1 of BSAcYNV has a non-AUG codon (UCUCUGG), it is in a suboptimal initiation context (the C of the CUG codon is designated +1, and there is pyrimidine instead of purine at -3 and a G at position +4) [12]. However, the ORF1 of BSAcVNV and the counterpart of BSMyV don't begin with a conventional AUG start codon as well, but they are both in an optimal initiation context (ACUCUGG and AUUCUGG corresponding to BSAcVNV and BSMyV respectively) with a purine at position -3 and G at position +4. The non-AUG start codon, in a context favourable for binding which is in accordance with the Kozak consensus sequence, could partially recognized by shunting ribosomes, as has been shown for Cauliflower mosaic virus [4]. But ORF 1 of BSOlV has a conventional start codon in a suboptimal initiation context and the AUG start codon for BSGfV ORF 1 lies in a very weak context (lacking both purines in position -3 and G in position +4), which is in agreement with the scanning model. The different context of sequence of the start codons for ORF1 of these BSV isolates might give rise to the variable abilities of ribosome recognition, as presented previously [6, 20]. It suggests the translational level of ORF1 genes may be diverse among these BSV isolates.
The ORF2 encodes a virion-associated protein (VAP) containing a coiled-coil domain which mediate not only homomerization but also interaction between heterologous proteins [17], and which is well-conserved in the plant pararetroviruses (Fig. 4). Alignment with ORF 2 of Badnaviruses revealed the same conserved coiled-coil domains. The coiled-coil domains of VAP can assemble into a parallel tetramer, a stable form in infected plants [21]. The VAP could act as the "arm" of the virus particle, attaching to the capsid shell by its C terminus [21]; and VAP might participate in cell-to-cell and long-distance movement via interaction with other proteins (e.g. viral MP), as shown in the Cauliflower mosaic virus [15].

Figure 4. Amino acid sequence alignment of BSAcYNV, BSAcVNV, BSOEV and BSMyV, and Commelina yellow mottle virus (ComYMV), Dioscorea bacilliform virus (DBV), Kalanchoe top-spotting virus (KTSV) ORF2 proteins. Asterisks denote exact matches, and double or single dots denote positions of conserved or semiconserved amino acid residues respectively. boxes indicate the conserved coiled-coil domains in Badnaviruses.
The ORF 3 encodes a 217.7 kDa polyprotein, which contains the viral movement protein (MP), the coat protein (CP), putative viral aspartic protease, reverse transcriptase (RT) and RNase H [2, 10]. MP and CP are characterized by a coiled-coil motif [YVIPDIMMT IRDFYRHIQI, the hydrophobic amino acid (in bold) form the helix interface, while the others are hydrophilic polar residues.] and the cysteine rich, zinc finger like RNA-binding motif (CXCX2CX4HX4C). Additionally, this polyprotein contains another zinc-binding domain CXHX11CX2CX11CX2CX4CX2C (Fig. 5), which was found downstream of the first zinc finger motif. Although the tangible function of this zinc finger motif remains unclear, we speculated that it might be involved in package of virions due to their potential nucleotide-binding activity [10]. The candidate NLSs (Nucleus Location Sites) of BSAcYNV represent in the putative CP region are PRRPRK (487-492, positions of the ORF3) and KRK (761-763, positions of the ORF3), so the multifunctional CP is also thought to be associated with transportation of the viral genome into nucleus via an importin α-dependent pathway as described in RTBV [17].

Figure 5. Alignment of the two Cysteine-Histidine motifs (A, B) of other badnaviruses and Rice tungro bacilliform virus (RTBV). The Badnaviruses are Banana streak virus isolates BSOEV, BSMyV, BSMyV, BSGfV, BSAcVNV and BSAcYNV, Commelina yellow mottle virus (ComYMV), Cacao swollen shoot virus (CSSV), Citrus yellow mosaic virus (CYMV), Dioscorea bacilliform virus (DBV), Kalanchoe top-spotting virus (KTSV), Sugarcane bacilliform virus (SCBV), Tara bacilliform virus (TBV). An asterisk denotes an exact match, and double or single dots denote positions of conserved or semiconserved amino acid respectively.
-
Through analysis by NCBI Blast, the BSAcYNV DNA sequence is found to be 88% identical to BSAcVNV, and their polyprotein amino acid (aa) sequences of share 95.1% homology. But overall the amino acid sequences show low identity to those of other BSV isolates (Table 1). Phylogenetic analysis of the zinc finger motifs showed that BSAcYNV was most closely related to BSAcVNV (Fig. 6), consistent with the neighbour-joining phylogenetic tree based on RT and RNase H domains (the data not shown). BSAcYNV and other four isolates did cluster together in a phylogenetic tree as well as forming a single cluster with Kalanchoe top-spotting virus (KTSV). This supports the idea that these banana streak badnaviruses from hosts with divergent genetic background can be regarded as a distinct virus species [17].

Table 1. Amino acid sequence homologies among BSAcYNV, other banana streak badnaviruses and Kalanchoe top-spotting virus (KTSV)
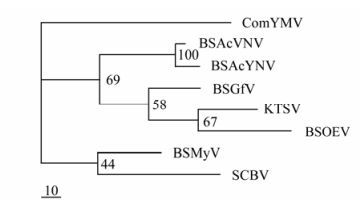
Figure 6. Phylogenetic tree constructed by the MrBayes program depicting the relationships of the Badnaviruses, Caulimoviruses and Rice tungro bacilliform virus (RTBV), based on the alignment of the reverse transcriptase and ribonuclease H domain. The badnaviruses are Banana streak virus isolates (BSOlV, BSMyV, BSGfV, BSAcVNV and BSAcYNV respectively), Commelina yellow mottle virus (ComYMV) Kalanchoe top-spotting virus (KTSV), Sugarcane bacilliform virus (SCBV).
BSV Purification and Genome Organization
Characterizations of ORFs
Phylogenetic analysis
-
The genome sequence of BSAcYNV demonstrates that this is a typical Badnavirus with a genomic structure similar to other badnaviruses. The pgRNA begins with a long leader, which carries a short ORF (sORF) in front of the ORF 1. This pgRNA leader sequence has been predicted to form an extended stem-loop structure. The structure brings into spatial proximity the sORF, consisting of six codons and which terminates 7nt upstream of the hairpin, and the ORF 1 located 23nt downstream of the hairpin (Fig. 5). The protein encoded by the sORF is highly conserved among BSV isolates [19]. The CU-rich stretch following the sORF stop codon might promote loosening of the contact between the RNA and the shunting ribosome, whereas the AU-rich region following the stable stem-loop is located in the spatial vicinity of a CU rich region; so the AU rich region might be involved in receiving the shunting ribosome [19]. It has been suggested the pgRNA leader sequence is bypassed by means of a ribosome-shunt mechanism, as proposed for CaMV and other plant pararetroviruses [11, 18]. Recently, in Drosophila, the SXL (Sex lethal) protein was found to bind downstream with poly U stretches of a short uORF (upstream open reading frame) that endows a negative effect on translation of the major reading frame, suggesting that the underlying regulatory principle may be broadly utilized in eukaryotic gene regulation [14]. Nevertheless, other RBPs (RNA binding proteins) similar to SXL have not yet been found in plants. Based on the ribosome-shunting model for translational initiation, the ribosome may shunt over the stable secondary structure and resume linear scanning on the downstream sequence of BSVs.
The pgRNA leaders of Banana streak viruses all possess an extensive stem-loop structure, whereas the dsRNAs region from the stem-loop structure may be cleaved into sRNAs (small RNAs) by a Dicer-like (DIL) protein; this mechanism has been observed in the 35S RNA leader of Cauliflower mosaic virus as a major source of CaMV-derived siRNAs and these siRNAs molecule potentially down-regulated host transcripts during infection [15]. Interestingly, the 35S RNA leader is well-conserved in sequence and structure among CaMV strains [15]; on the contrary, the pgRNA leaders from different BSVs species, given the greater variation in their sequence, appear to have evolved to produce a diverse range of secondary structures. Also, discrete vsRNAs (virus-derived small RNAs) from several mammalian DNA viruses have been shown to regulate the viral genome expression in cis and inhibit expression of specific host gene transcripts [7, 22]. This might be reminiscent of the extensive stem-loop structures that exist in BSV which act as sources of virus-derived siRNAs, and which are thought to guide cleavage of viral transcripts after their incorporation into a RNA-induced silencing complex (RISC). The variation in vsRNA properties originated from the different pgRNA leaders at transcriptional level could be attributed to differences of interfering regulatory roles in viral infections on different phenotypes banana hosts which correspond to distinct BSV species. If this were the case, this would bring about production of truncated BSV pgRNA transcript and the deletion transcript might be reverse transcribed into defective DNA (in contrast to the complete viral genome), as subgenomic DNA of progeny virions. This proposed model may explain the occurrence of defective genomes in BSAcYNV. However, it cannot be excluded that other possiblities (such as homologous recombination, integration etc.) could also contribute to the occurrence of these sub-genome sequences. To date, whether the defective genomic DNA of BSAcYNV is a discrete replicon in infected hosts has not been determined.
Among the all BSV isolates, the highest identities at both nucleotide and aa levels were shared by BSAcYNV and BSAcVNV; moreover, the neighbor-joining phylogenetic trees suggested BSAcYNV has the closest evolutionary relationship to BSAcYNV in banana streak badnaviruses. Given their adjacent geographical regions (BSAcYNV is from Yunnan province in China, BSAcVNV is from Vietnam, and Yunnan province borders Vietnam.) and their hosts with similar genetic background (the hosts of the two BSV isolates were both isolataed from M. Acuminata), BSAcYNV and BSAcVNV may be regarded as close virus isolates that can be discriminated from other distinct BSV strains.













 DownLoad:
DownLoad: