-
Bacterial strains with surface expression of viral proteins can be used for the development of a diagnostic system for viral diseases or a live oral vaccine against those diseases [11-13], and for the studies of the interaction between virus and host by simulation [2, 15]. Many bacterial cell surface localized molecules can act as display motifs to target heterologous proteins on their surface. Among those, an ice nucleation protein (INP) derived from Pseudomonas syringae has been extensively used as a display motif in bacteria. INPs can stably display proteins with molecular weight as high as 90 kDa (Chi92) and 60 kDa (gp120) in Escherchia coli and Salmonella species [12, 20]. The displayed proteins can be easily verified [11] and show no evidence of interference in the structure or function of the bacterial cell membrane [18].
INP contains three domains: an N-terminal unique region composed of 191 amino acids, a central repeating domain (CRD) consisting of a series of repeated amino acid sequences with lengths of 8-, 16-, and 48-residues which act as templates for ice crystal formation, and a C-terminal unique region of 49 amino acids. The N-terminal domain (INPN) can be anchored to the membrane, and the C-terminal region (INPC) is highly hydrophilic and exposed to the outermost cell surface [7-9]. There are several strategies to display the foreign proteins using this system. The foreign protein can be inserted into CRD directly, or fused with the N-terminal domain with a depletion of CRD and C-terminal domain, or fused with N-and C-terminal domain without CRD (INPNC). Because of the size limitation for detection of foreign proteins, the full-length INP is not applicable in some situations; foreign proteins with high molecular weights can be displayed using INPN [12, 20], but the addition of INPC may improve the stability of fusion protein [14].
S. typhimurium is a gram-negative, facultative intracellular bacterium, and belongs to O-group B [6]. Due to their ubiquitous ability to invade eukaryotes, S. typhimurium and the attenuated strains are generally used as carriers to transfer a plasmid into the host cell and to elicit immunity against passenger antigens [4]. Virus antigen presentation can be enhanced by a surface-displayed mechanism: a fusion protein encompassing a dengue virus epitope can reach the cytosol of CTLs through destabilizing the membrane of a phagosome containing surface-displayed Salmonella, then the virus epitope can be processed through the MHC class I-dependent pathway to stimulate CTLs similar to a living virus [15]. However, there remains uncertainty in using INP-based cell surface display with Salmonella strains, in particular there are concerns regarding the integrity of foreign protein and the display efficiency of different display motifs. S. typhimurium BRD509 is a mutant (aroA, aroD) derived from virulent S. typhimurium SL1344 [17], containing plasmids that appear to be quite stable [3].
In this study, the displays of enhanced fluorescent green protein (EGFP) and domain Ⅲ of Japanese encephalitis Virus (JEV) E protein (JEDIII), with or without EGFP tag, on the surface of S. typhimurium BRD509 using INPN and INPNC were investigated. In comparing the integrity among the displayed proteins, some characteristics of the use of the INP-based cell surface display system in Salmonella strains were revealed. Our findings may give a better understanding of the mechanism of ice nucleation protein in displaying foreign protein and developing an effective live vaccine against viral disease.
HTML
-
Plasmid pTrcHisC (Invitrogen) or pGEM-T easy (Promega) was used as cloning vector and pTrchisC was also used as expression vector. Plasmid pMPL003 (a kind gift from Dr. Lin Li, Huazhong Agricultural University, China) containing the full-length gene inaK of P. syringae KCTC1832, plasmid pEGFP-N1 (Clonetech) containing egfp encoding full-length EGFP and plasmid pCJEVprME (unpublished) containing full-length gene pr-Membrane and the E gene of JEV (Beijing P3 strain) were used as templates for gene cloning. E. coli TOP10 (also a kind gift from Dr. Lin Li) was used as a host for cloning, protein expression and display of JEDIII. S. typhimurium BRD509 (a kind gift from Derek Pickard, Wellcome Trust Sanger Institute, UK) was used as a host for surface display of JEDIII, or EGFP. Both bacterial strains were cultured in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 37℃ or 25℃, and 50 μg/mL ampicillin was supplemented at final concentration for bacteria containing plasmid. African green monkey kidney (Vero) cells were derived from ECACC and passaged at the Brain Infections Group of University of Liverpool. Vero cells were grown in RPMI1640 (Gibco) supplemented with 5% fetal calf serum, 100 μg/mL penicillin and 100 μg/mL streptomycin at 37 ℃ at humidified atmosphere with 5% CO2.
-
All primers are listed in Table 1 and plasmid construction is illustrated in Fig. 1. By using primer pairs EGFP-F/EGFP-R and INPN-F/INPN-R, gene egfp and gene encoding INPN (aa residues 1-191) were amplified from plasmids pEGFP-N1 and pMPL003 by PCR, respectively. The PCR products digested by restriction enzymes PstⅠ/KpnⅠ and XhoⅠ/PstⅠ were inserted into the same enzyme-digested pTrcHisC, resulting in recombinant plasmids pTEGFP and pTINPN, respectively. By using primer pair JEDIII-F/JEDIII-R, gene encoding JEDIII (aa residues 292-402 of JEV E protein) were amplified from plasmid pCJEVprME by PCR. The PCR products were inserted into pTrcHisC at restriction enzyme sites of XhoⅠ and KpnⅠ, giving recombinant plasmid pTrcJEDIII. Furthermore, the gene encoding INPN (aa residues 1-191) amplified by primers InpN-O-F/InpN-O-R was linked with amplified products from gene encoding INPC (without stop codon) using primers InpC-O-F/InpC-O-R by Overlapping PCR. The fusion gene was sub-cloned into pGEMT-easy at the restriction sites XhoⅠ and PstⅠ to obtain recombinant plasmid pGEMTINC.
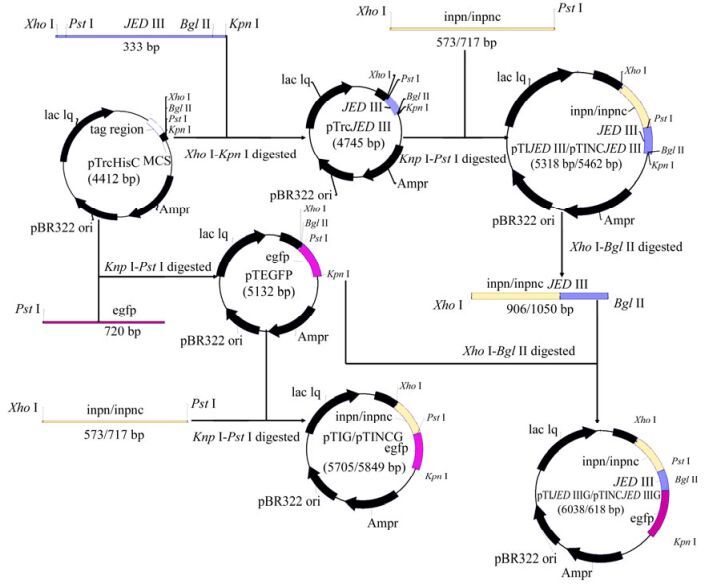
Figure 1. Schematic diagram of construction of plasmids pTrcJEDIII, pTEGFP, pTIJEDIII, pTINCJEDIII, pTIG, pTINCG, pTIJEDIIIG and pTINCJEDIIIG.

Table 1. Primers used in this study
The 573 bp XhoⅠ-PstⅠ digested fragment from pTINPN was inserted into the same enzyme-digested pTrcJEDIII and pTEGFP, resulting in recombinant plasmids pTIJEDIII and pTIG, which contain a fusion gene encoding INPN-JEDIII (predicted size: 37 kDa) and a fusion gene encoding INPN-EGFP (predicted size: 51 kDa), respectively; The 717 bp XhoⅠ-PstⅠ digested fragment from pGEMTINC was inserted into the same enzyme-digested pTrcJEDIII and pTEGFP to obtain pTINCJEDIII and pTINCG, containing a fusion gene encoding INPNC-JEDIII (predicted size: 42 kDa) and a fusion gene encoding INPNC-EGFP (predicted size: 56 kDa), respectively.; The 906 bp XhoⅠ-BglⅡ digested fragment from pTIJEDIII and the 1, 050 bpXhoⅠ-BglⅡ digested fragment from pTINCJEDIII were inserted into the same enzyme-digested pTEGFP to obtain pTIJEDIIIG and pTINCJEDIIIG, containing a fusion gene encoding INPN-JEDIII-EGFP (predicted size: 64 kDa) and a fusion gene encoding INPNC-JEDIII-EGFP (predicted size: 69 kDa), respectively.
Recombinant plasmids were electroporated into E.coli TOP10 and S. typhimurium BRD509 after being verified by DNA sequencing. Recombinant bacterial strains were designated by the name of their containing plasmid additive with prefix "B-" (Parental strain was S. typhimurium BRD509) or prefix "T-" (Parental strain was E.coli TOP10). For example, B-pTIJEDIII denoted S. typhimurium BRD509 containing plasmid pTIJEDIII.
-
The recombinant bacterial cells were used to prepare a cytoplasmic fraction (CP) and an outer membrane fraction (OM) according to methods described elsewhere [14, 19] after being induced upon 0.1 mmol/L IPTG at 25 ℃ for 24 h. Fractionated samples were resolved by 12%(wt/vol) (samples containing protein with predicted size larger than 60 kDa) or 10% (wt/vol) SDS-PAGE, followed by electrophoretic transfer to Hybond-PVDF membranes (Amersham Pharmacia Biotech, UK). For Western blot analysis, rabbit anti-JEDIII polyclonal sera (prepared by our group), or mouse monoclonal antibody anti-Xpress™ (Invitrogen), or mouse monoclonal antibody GFP3E1 (a kind gift from Dr. Neil Blake, University of Liverpool, UK) was used as primary detecting antibody. One-Step™ TMB-Blotting Substrate (KPL) was used as a substrate against peroxidase conjugated with the secondary antibody. Membranes were photographed and analyzed using the GeneSnap software provided with the Chemigenius Bioimaging System (Syngene) after color developing.
-
Vero cells were cultured in glass coverslips (pretreated 6-well Corning plates). Recombinant B-pTIJEDIII was freshly prepared for invasion assays through induction by 0.1 mmol/L IPTG at 25 ℃ for 24 h. Vero cells were infected at a multiplicity of infection (MOI) of 20: 1 (CFU of bacteria: count of Vero cells) for 2 h at 37 ℃ in the presence of 5% CO2. After 2 h incubation, cells were washed three times with PBS containing 100 µg/mL gentamicin to remove extracellular bacteria, and then coverslips were removed to be used in immunohistochemical staining [1, 4, 5].
-
Culture cells on coverslips were fixed using methanol at -20 ℃ for 10 min, blocked with 10% (vol/vol) normal goat serum in TBS buffer (20 mmol/L Tris-HCl, 500 mmol/L NaCl, pH7.5) at room temperature(RT) for 1.5 h, and then permeabilized using 0.1% (vol/vol) Triton X-100 (Sigma Aldrich) at RT for 4 min. Subsequently, preparations were incubated in anti-O4 rabbit polyclonal serum (1: 3 000 (vol/vol) (Remel Europe Ltd, Dartford, UK) or Anti-Xpress™(1: 5000, vol/vol) at RT for 1 h, washed three times with TBS for 5 min, and then incubated in anti-Rabbit IgG conjugated peroxidase (1:5000, vol/vol) or anti-mouse IgG conjugated alkaline phosphatase (1: 5000, vol/vol, Sigma) at RT for 1 h. The washing procedure was repeated with TBS, and after the addition of one-Step™ TMB-Blotting Substrate (KPL) or BCIP/NBT (Sigma-Aldrich) the cells were incubated at RT for 30 min. After quenching with distilled water, coverslips were counterstained with 1% eosin at RT for 4 min, rinsed with tap water, and then mounted by Verta Mounting Media (Sigma-Aldrich). Images were captured using a digital camera and and analyzed using imaging software (Nikon ACT-2U) [1, 5].
Strains B-pTIJEDIII, T-pTIJEDIII, S. typhimurium BRD509 and E. coli TOP10 were induced with 0.1 mmol/L IPTG at 25 ℃ for 24 h. Harvested cells were then washed four times with TBS buffer (20 mmol/L Tris-HCl, 500 mmol/L NaCl, pH 7.5). Cells were blocked at RT 2 h in TBS containing 3% BSA, and then incubated in solution containing Anti-Xpress™ (1: 5 000, vol/vol) at RT 1.5 h. After washing three times, the cells were incubated in secondary anti-mouse antibody conjugated alkaline phosphatase solution for 2 h, washed again, and then incubated in BCIP/NBT solution for 10 min at RT. After quenching with distilled water, photos were taken by digital camera [14].
Cell lines, Bacterial strains, Plasmids and Culture Conditions
Plasmid Construction
Western Blot Analysis
Invasion assay
Immunohistochemical staining
-
SDS-PAGE and Western blot analysis demonstrated that recombinant strains B-pTIJEDIII and B-pTINCJEDIII could produce 37 kDa and 42 kDa fusion proteins after induction, respectively, and proteins with the predicted size were detected in the OM from recombinant strains either by using Anti-Xpress™ (Fig. 2a) or anti-JEDIII(Fig. 2b) as the detecting antibody; A 51 kDa protein was detected in the OM from B-pTIG by using GFP3E1 as the detecting antibody and a 56 kDa protein from B-pTINCG was detected by using GFP3E1 or Anti-Xpress™ as the detecting antibody, indicating that JEDIII and EGFP were expressed on the surface of S. typhimurium BRD509. Although the green fluorescence of recombinant S. typhimurium containing pTINCJEDIIIG or pTIJEDIIIG can be seen by naked eye, violet analyzer or fluorescence microscope after being induced upon IPTG, no protein bands with the predicted size were detected in the OM from the recombinants (Fig. 2c and d). The results revealed that JEDIII with EGFP tag cannot be functionally expressed on the surface of S. typhimurium BRD509 or that the fusion proteins could be expressed but underwent proteolysis.

Figure 2. Western blot analysis for cell fraction of Salmonella typhimurium BRD509 harboring recombinant plasmids. Bacteria were cultured in LB at 25℃ for 24h induced with 0.1mmol/L IPTG. CP, cytoplasmic fraction; OM, outer membrane fraction. M, ProtoMarkers™ Pre-stained Protein Standards; EGFP, enhanced green fluroscence protein; JEDIII, purified protein Domain Ⅲ of JEV E protein; OM, outer membrane fraction; CP, cytoplasmic fraction. B-pTINCG, S. typhimurium BRD509 harboring pTINCG; B-pTIG, S. typhimurium BRD509 harboring pTIG; B-pTINCJEDIIIG, S. typhimurium BRD509 harboring pTINCJEDIIIG; B-pTIJEDIIIG, S. typhimurium BRD509 harboring pTIJEDIIIG; B-pTIJEDIII, S. typhimurium BRD509 harboring pTIJEDIII; B-pTINCJEDIII, S. typhimurium BRD509 harboring pTINCJEDIII; . (a) using Anti-Xpress™ for detection; (b) using anti-JEDIII for detection; (c) using GFP3E1 for detection; (d) using Anti-Xpress™ for detection.
-
Immunohistochemical studies showed that that the intact B-pTIJEDIII and T-pTIJEDIII could be blue-stained (positive) after being induced upon IPTG; whereas blank Salmonella and E. coli were unstained (Fig. 3). Along with the results of Western blot analysis mentioned above, this indicated that INPN can target JEDIII onto the surface of S. typhimurium BRD509 just as it did in E. coli TOP10. As in other studies [14], INPN can be used as a motif to display foreign proteins onto the surface of S. typhimurium alone.
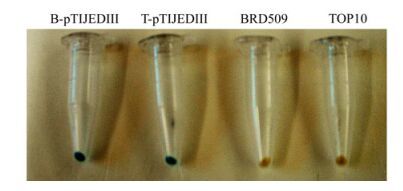
Figure 3. Immunohistochemical staining of the intact cells of B-pTIJEDIII and T-pTIJEDIII. Bacteria were cultured in LB at 25℃ for 24h induced with 0.1mmol/L IPTG, then analyzed by immunohistochemical staining, using Anti-Xpress™ as detecting antibody.
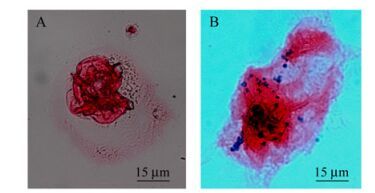
Invasion assay and immunohistochemical staining showed that the B-pTIJEDIII could infect Vero cells (Fig. 4). It indicates that Salmonella with surface-expressing JEDIII retained their invasiveness; thereby, this recombinant strain can be used as a live vaccine against viral infection. Because the integral protein has complete immunogenicity, integrity of JEDIII on the bacterial cell surface needed to be verified.
-
All recombinant plasmids were constructed from parental plasmid-pTrcHisC, which has a sequence encoding an N-terminal fusion peptide, including six histidine residues and an Xpress™ epitope, thus, the integrity of the fusion protein can be confirmed by using antibody (Anti-Xpress™) against the Xpress™ epitope along with antibody (anti-JEDIII or GFP3E1) against passenger proteins (JEDIII or EGFP) in Western blot analysis.
Protein bands with the predicted size can be detected in the OM and the CP from B-pTIJEDIII or B-pTINCJEDIII, using Anti-Xpress™ (Fig. 2a) or anti-JEDIII (Fig. 2b). Furthermore, only one protein band is shown in the OM from those two strains. This indicates that the fusion protein INPN-JEDIII or INPNC-JEDIII can be completely expressed on the bacterial cell surface. Interestingly, a protein band with almost the same size as integral INPN-JEDIII can be found in the CP (Fig. 2b) using anti-JEDIII as detecting antibody, whereas no protein band can be found (Fig. 2a) when using Anti-Xpress™ for detection. It seems that the Xpress™ epitope underwent functional disruption instead of physically degrading, and it implies that the degradation of fusion protein INPN-JEDIII was perhaps initiated from its N-terminus while targeting onto the membrane from the cytoplasm. However, no similar results were shown when JEDIII was displayed on S. typhimurium, using INPNC as display motif, implying that using INPNC to display JEDIII is very stable. Nevertheless, the signal of INPNC-JEDIII was very weak in Western blot analysis, compared with that of INPN-JEDIII at the same loading of their fractionated samples. This may be ascribed to lower display efficiency for INPNC.
More than one protein band can be seen in the OM and CP from B-pTIJEDIIIG or B-pTINCJEDIIIG by Western blot analysis, using GFP3E1 as detecting antibody (Fig. 2c), whereas no protein bands can be detected by Anti-Xpress™, either in the OM or CP (Fig. 2d). This shows that fusion protein INPN-JEDIII-EGFP and INPNC-JEDIII-EGFP cannot be correctly expressed in Salmonella. Therefore comparison of the relative display efficiencies of INPNC and INPN cannot be performed by measuring fluorescence intensities of tagged EGFP. However, as mentioned above, there was only one protein band in OM from B-pTIJEDIII or B-pTINCJEDIII, so sandwiched-ELISA can be used to measure the content of fusion proteins in the OM. Also, Triton X-100, used to prepare the OM in our study, was reported to compete with antigen to bind antibody [10], thus, it can interfere with the result of ELISA.
Though more than one protein band can be seen in the OM or CP from B-pTINCG and B-pTIG, using GFP3E1 as detecting antibody, protein bands of the predicted size can be found (Fig. 2c); whereas only one protein band with the predicted size in OM from B-pTINCG can be detected using Anti-Xpress™ (Fig. 2d). Those results were quite different from a similar study in E. coli; fusion protein INPNC-GFP showed no degradation in the OM of E. coli when GFP was displayed by using INPNC, whereas free GFP could be detected in the OM when GFP was displayed using INPN [14]. This may be ascribed to different membrane-associated protease distributions between Salmonella strains and E. coli strains [16]. For practical application, the efficiency of INP displaying foreign protein on S. typhimurium should be further investigated.
Both display motifs from INP can target JEDIII or EGFP onto the surface of S. typhimurium, and INPC can improve stability of fusion proteins encompassed by JEDIII or EGFP in the outer membrane, whereas INPN can target integral JEDIII onto bacterial surface alone. These findings can improve understanding of the mechanism of cell surface display in S. typhimurium by using INP.













 DownLoad:
DownLoad: