-
Rabies virus (RABV) belongs to the Mononegavirale order, the Rhabdoviridae family and the Lyssavirus genus. The genome is a non-segmented, negative-sense, single-strand RNA which is about 12, 000 nucleotides (nt) long. This viral RNA encodes five structural proteins in order: Nucleoprotein (NP), phosphoprotein (PP), matrix protein (MP), glycoprotein (GP) and large protein (LP).
Sequencing and analysis on the complete genome of rabies virus were started in 1988 [20], and more than 70 strains of rabies viruses have been subjected to complete genome sequencing according to NCBI index till 2011. The complete genome sequencing of the rabies virus has important significance for completely understanding molecular evolution, gene regulation and genetic variation of the rabies virus.
China is a country with high morbidity for rabies and ranks the second highest place in the world after India. Rabies also leads in the number of reported deaths among the various kinds of communicable diseases of the Notifiable Communicable Disease Reporting System in China. The reported cases mainly occur in south China, and among them more than 90% of the cases are found in Yunnan, Guangdong, Guangxi, Hunan and other provinces or autonomous regions [23]. It should be noticed in these areas with high morbidity, such as in Yunnan province, the reported human rabies cases(The data was collected as part of the "National Disease Reporting Information System" of the Chinese Center for Disease Control and Prevention) has rapidly increased and expanded in recent years (Fig. 1). Only one or few cases was reported in 2000, 2002 and 2003 respectively in Yunnan, and rabies showed scattered prevalence before 2005 but it rapidly increased in 2006, 108 cases were reported in 2008 and 134 cases were reported in 2010.
Yunnan province with its special geographic location is adjacent to Tibet, Sichuan, Guizhou and Guangxi provinces of which Sichuan, Guizhou and Guangxi are areas with a high incidence of rabies. It is also adjacent to Laos, Burma, Vietnam, and close to Thailand where rabies is prevalent.
In recent years, the full-length genomes of some vaccine and street strains of rabies viruses were determined [3, 13, 16, 18], which establish the foundation for understanding the characteristics of hereditary variation for rabies virus in China. In this study, we isolated and sequenced a street strain of rabies virus at the borderline area of Yunnan, with an aim to explore the correlation between the rapid increase and expanding of rabies in Yunnan and the hereditary variation of rabies virus.
HTML
-
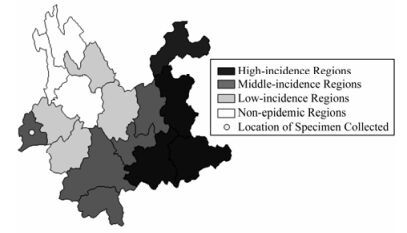
The CYN1009D strain was isolated from the brain tissue of a rabid dog in Lianghe county, Dehong Prefecture, Yunnan province(as shown in Fig. 2), and subjected to 1 passage in the suckling mice to amplify the virus. The brain tissue of the moribund suckling mice was used to isolate the total RNA.
-
In total, 16 pairs of primers spanning the entire CYN1009D genome were designed using Primer Premier, version 5 (PREMIER Biosoft International, CA, USA), and all primers were designed within the conserved regions of the genome, determined from an alignment of all the full genomic sequences of the reference strains (Table 1).

Table 1. Primers for amplification of the whole genome of CYN1009D strain
-
Total RNA was extracted from brains of mice infected with the CYN1009D strain using TRIzol reagent (Invitrogen, Carlsbad, CA). Single-strand cDNAs were synthesized by reverse transcription using Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions. Briefly, 32 μL of total RNA was heated at 60℃ for 10min, then quickly chilled on ice for at least 2 min, and then transferred to a reaction tube of Ready-To-Go You-Prime First-Strand Beads with 1μL of random primer pd(N)6 (0.2 g/mL) (TaKaRa, Japan). After incubation at 37℃ for 60 min, the synthesized cDNA product was used for PCR.
-
16 overlapping fragments were amplified and the primers are shown in Table 1. The cycling parameters were one cycle at 94℃ for 3min for an initial denaturation, then 35 cycles at 94℃ for 30 s, 52-57℃ for 30 s, and 72℃ for 80 s, and a final extention at 72℃ for 10 min. The PCR products were identified on 1% Agarose (Invitrogen, USA) gel and visualized by ethidium bromide staining under UV illumination with 2 kb DNA markers (TaKaRa, Japan). All of the PCR products of the expected ED PROOF size were then excised from the agarose gel and purified with the QIAquick Gel Extraction Kit(Qiagen, Germany), following the manufacturer's instructions. The purified products were then directly sequenced commercially (Shanghai Sangon Biological Engineering, China).
-
To determine the sequence at the 3' end of RABV genome, PCR amplification was carried out according to the previous method [14]. Briefly, 4 uL (80 pmol/L) 5' phosphorylated DNA oligonucleotide 3RACE primer P (5'-P-GTCGATCACGCGATCGAACGGTCGCTGAG-3') [12], 4 uL RNA, 5 uL T4 RNA Ligase Buffer, 3uL 0.1% BSA, 1 uL T4 RNA Ligase (50 U/L), and 36 uL DEPC treated water were successively added in the tube, and then incubated at 12℃ for 16h, 75% ethanol was used to precipitate the ligation product and subsequently 10uL DEPC treated water was added. 20 pmol oligonucleotide 3RACE inner primer (5'-CAGCGACCGTTCGATCGC-3') was used for cDNA synthesis. 3RACE inner primer and 3GSP (5'-CGACCATCATCAGGATCAAG-3') were to gen erate an amplicon of 290 bp. To determine the PCR product at the 5' end of RABV, a 5' Full RACE Kit was used in the present study according to the manufacturer's instructions. The primers were as follows: 5' GSP1: 5'-CGATGAAGCAGGTTATTCG AGGGA-3', 5'GSP2: 5'-CGATCTCTAGCTTGAGT CTGTC-3'.
-
The resulting sequences were assembled and manually checked using the ATGC program version 4 (Genetyx Co., Tokyo, Japan). The ClustalX version 1.83 was used for multiple alignments of the sequences. DNAStar (version 5.01) was used to translate the gene sequences and to determine the percentage identities and similarity scores. Phylogenetic trees were constructed with MEGA, version 5. The background data for the rabies virus in this study are shown in Table 2.

Table 2. The genome sequences of rabies viruses used in this study
Virus strain
Primer design
RNA isolation and reverse transcription
PCR and sequencing
Amplification for 3' end and 5' end of RABV genome
Sequence alignment and analysis
-
The full length of the CYN1009D genome (GenBank accession no. JQ730682) strain is 11923 nt, the genomic organization of CYN1009D, which is typical of all previously characterized RABVs, is as follows: 3' leader region (1-58), N gene (59-1483), P gene (1486-2474), M gene (2480-3282), G gene (3288-5354), L gene (5379-11853) and a trailer region (11854-11923). All five genes are initiated with AACA and terminated with poly (A)7. The coding sequences (CDS) of each of the five structural genes are located as follows: N gene 1353nt (71-1423), P gene 894nt (1514-2407), M gene 609nt (2495-3103), G gene 1575nt (3315-4889), L gene 6387nt (5406-11792); The monocistrons are separated by intergenic regions of 2, 5, 5 and 24 nucleotides respectively.
-
The N gene of the CYN1009D strain encodes a protein of 450 amino acids residues, which are identical to the lengths of other strains in China. In comparison to other wild strains in China, the antigen site Ⅰ (358-367) and antigen site Ⅲ (313-367) of CYN1009D are completely conserved, variation was only detected at one position in the antigen site Ⅳ (359-383) Thr375→Met375. Furthermore, CYN1009D is different from other strains in China at five sites: Val111, Val128, Ala135, Val379 and Ala426, Thr42, which are identical to those in SRV9, GX4, HN10 and CTN-181, but different from those in FJ008, FJ009, D01 and SH06. Among them, the hydrophilic amino acid Ser(S) at site 135、426 is replaced by hydro-phobic amino acid Ala(A), other variation points are between the same type of amino acid variation. Other structural and functions sites are all conserved.
PP is the structural protein with the most significant variations in the rabies virus. The P gene of CYN1009D strain encodes a protein of 297 amino acids residues, which is identical to other strains in China in its length. Amino acids from 1-19 encode the synergetic factor of LP which participates in transcription and replication of viral RNA. It is the major site for the action of PP and LP [1] and is highly conservative in CYN1009D. The amino acids from 143 to 148 are the site of PP and the light chain LC8 of the cytoplasmic dynein, and variation was not detected in this region in CYN1009D strain and they were all DKSTQT [8]. The amino acids FSKKYKFP from 209 to 216 are major areas for the interaction between PP and NP [7], and this region is also highly conservative in CYN1009D. Amino acids at 63~64 as well as 162, 210 and 271 have specific serine sites for protein kinase phosphorylation, which can phosphorylate PP under the effects of protein kinases, and they are major phosphorylation sites in the P gene [6]. In comparison to other strains in China, CYN1009D is conservative in amino acids at 210 and 271, but variations were detected at 63-64 and 162. Furthermore, there are more than 20 variation sites in CYN1009D.
MP is a protein that has completely identical length in all of the genotypes of rabies viruses, and The M gene of CYN1009D strain encodes a protein of 202 amino acid residues. The PPXY domain playing an important role in viral budding (35-38aa) is highly conserved. The special domain at 35~38 in MP of CYN1009D PPxY is PPEY. MP can negatively control viral transcription, but its regulatory sites during viral transcription and replication are not identical to those during viral assembly and budding [5], and the amino acid at position 58 in the site of negative control on transcription was E in CYN1009D, which is identical to the strains in China except SRV9 (R).
The G gene of CYN1009D strain encodes a protein of 524 amino acids residues, which is identical to other strains in China. GP is the only glycosylated protein among the structural proteins in the rabies virus, and the sites and number of glycosylation in different strains are different [22]. The glycosylation motif of the G protein is the Asn-X-Thr/Thr motif, while CYN1009D has only two common glycosylation sites: Asn37 and Asn319. Among other strains in China, position 247 in SRV9 and CTN-181 is NET and it is DET/DEI in other strains, which is different from that in CYN1009D (DEA). The neutralizing antigen site Ⅰ (231), Ⅱ (34-42 and 198-200), positions Ⅲ (330-338) and a (342-343) are conserved. Amino acids at 333, 336, 339 and 357 having the closest correlation to antigenicity [19] are conserved. In comparison to the wild strains in China, the transmembrane region of GP shows significant variations (440-461), the five amino acids are completely different from those in the wild strains in China, and two amino acids are the same as those in some strains.
The L gene of CYN1009D strain encodes a protein of 2128 amino acids residues. There are six conservative regions in the structure of LP [15, 17] (which is determined by the positions of amino acids in PV strain): Ⅰ (232-423), Ⅱ (504-607), Ⅲ (608-831), Ⅳ (889-1060), Ⅴ (1090-1326) and Ⅵ (1673-1747); the short sequences (528-DLGDLPD-534 and 582-DALTMD-588) which are similar to the binding domains for magnesium ion (DXXXXXD) [21] in the conservative Ⅱ region are highly conserved; Asp1706 is extremely conserved in rabies viruses and this site may affect the methyltransferase activity of L protein, and it is also conservative in CYN1009D. The unique GDNQ motif for forming phosphodiester bond in the rabies virus can be found in the conservative sequence Ⅲ region of LP, and it is also termed as the RdRp active center motif. Any change in this element may lead to complete loss of polymerase activity [11], and this motif is absolutely conserved in all of the rabies virus strains used for comparison.
Excepting only one amino acid which was lost in SRV9 among the strains in China, the other strains are all composed of 2128 amino acids. The differences are found at the starting position of LP (Fig. 3), SRV9 starts with an M, while the other strains all start with two continuous M, but CYN1009D starts with ML, which is different from other strains in China. However, L and M are both nonpolar amino acids.
-
The composition of nucleotides in the intergenic regions of rabies viruses differs significantly. It can be found from the analysis in Table 3 that the N-P intergenic regions are all CT except that it is CG in the FJ009 street strain and TT in the F02 street strain. The P-M intergenic regions and the M-G intergenic regions for most strains are CAGGC and CTATT, while the third nucleotide in the M-G intergenic regions in CYN1009D, F02, GX4, CTN-181 and HN10 show a variation from A→T. The G-L intergenic region is the longest and shows the most significant variation.

Table 3. Intergenic region analysis between CYN1009D and other stains in China.
-
3'UTR and 5'UTR in the RABV genome plays regulatory roles in viral transcription and replication [4]. The 3' UTR of CYN1009D strain is composed of 58 nucleotides and its 5'UTR is composed of 70 nucleotides. Among them 11 nucleotides in 3'UTR and 5'UTR of CYN1009D strain are conservative and completely reverse complementary.
-
Whole-genome phylogeny (Fig. 4) using nucleotide sequences reveals that CYN1009D and the street strains in China are distributed in two different clusters, while it is allocated in the same cluster with 8764THA and 8743THA from Thailand, indicating that they have a closer genetic relationship.
GP is the major determinative factor for pathogenesis of viruses and the G gene is significantly affected by selection pressure, which is always related to highly synonymous mutation and is an ideal target for investigations on the natural evolution of viruses [10, 24]. The present study selected the G genes from other strains from Yunnan province, strains from other provinces adjacent to Yunnan and representative strains from other Asian countries to construct the cladogram. The results show that the strains in Yunnan province can be divided into two clusters, some strains in Yunnan were allocated in the same cluster with the strains from Guangxi, Fujian, Zhejiang and Chongqing, indicating that they have a relatively close genetic relationship; while the CYN1009D strain and another strain from Yunnan are allocated in the same cluster with the strains from Southeast Asian countries such as Burma, Laos, Vietnam, Cambodia, Thailand and Malaysia. The strains from other Asian countries are allocated in another two clusters.
The genomic organization of the CYN1009D isolate
Structural proteins' features of CYN1009D
Intergenic region analysis
Nucleotide sequence analysis for the terminal regulatory sequence
Phylogenetic analysis
-
The complete genome of CYN1009D is composed of 11923 nt and it belongs to genotype Ⅰ, which did not significantly differ from the complete genomic sequences of other rabies viruses published in GenBank, but is identical in length to a previously determined strain by our laboratory HN10. Among the genotype Ⅰ of Lyssavirus, the lengths of complete genomes are normally between 11923 nt and 11928 nt [2], a strain with 11922 nt (GX4) has also been reported in China, but the significance for the length of the complete genome for the characteristics of viruses is still unknown.
In the comparisons in major functional sites between CYN1009D and other strains, particularly the antigen sites of NP and GP, mutations in some amino acids were detected. However, mutations were not detected in key positions, such as Ser89 of NP which is related to the capability for viral transcription and replication, PPxY of MP which plays an important role in viral budding, positions 330 and 333 of GP which are directly related to the virulence of viruses, which also indirectly implies the important roles of these sites. Thr375 in the antigen site IV (359-383) of NP is highly conservative among the vaccine strains and the wild strains, but it is mutated into Met in the CYN1009D strain Position 375 is a minor phosphorylation site and the phosphorylation at this site is affected by the phosphorylation at position 389 Once position 389 was dephosphorylated, position 375 was also dephosphorylated accordingly [9], and the effects for the mutation at this site on the strain still need further study.
It was found in previous studies that the LP transcription of some rabies viruses was started with two Met residues and they were relatively independent in phyletic evolution, which is mainly found in wild strains in China, and it has not been reported in the strains from France, India, Brazil and Thailand [13]. The second amino acid in CYN1009D in the present study is mutated into a nonpolar amino acid L. The functions of this mutation in virus evolution and transmission are still unclear now and more information can be discovered after further analysis is carried out on LP in rabies viruses from other areas.
The phylogenetic analysis of the G gene shows that the geographical characteristic of rabies virus in Yunnan is that the specimens from the same province (the same geographical location) tend to evolve to the same evolutionary cluster or sub-cluster. Some strains in Yunnan show relatively close genetic relationship to those in adjacent provinces such as Guangxi and Sichuan, while the CYN1009D strain shows relatively close genetic relationship to the strains from Southeast Asian countries particularly the strains from Burma, indicating that the epidemic strains of rabies virus in Yunnan have a close genetic relationship to the strains from adjacent provinces and adjacent countries in Southeast Asia. It is estimated that the prevalence of rabies in Yunnan is related to the distribution and expanding of rabies viruses, and the possibility for expanding from other countries can not be ruled out in addition to the general expansion from different provinces. The geographical expansion of the Chinese strains in China and simultaneous expansion in adjacent countries may affect the prevalence and distribution of rabies in China and adjacent countries. Therefore, the prevention and control rabies at borderland areas should be reinforced simultaneously when efforts are made to control rabies in China, which is under control. These are essential procedures for the effective prevention and control of rabies all over the world.
















 DownLoad:
DownLoad: