-
The initial step in cell infection by retroviruses, including lentiviruses occurs with the binding of envelope glycoproteins (Env) to target cells. Knowledge of the genetic variation and glycosylation modification of Env is important for understanding the molecular mechanisms underlying virus-cell interactions and for the design of effective vaccines to viruses. Human immunodeficiency virus (HIV), simian immunodeficiency virus (SIV), feline immunodeficiency virus (FIV) and equine infectious anemia virus (EIAV) are all members of the lentivirus genus of the retroviridae family, and infection by members of this group of viruses often lead to immunosuppression such as AIDS caused by HIV. The first and widely used lentivirus vaccine is the vaccine against EIAV, which was developed three decades ago [25].
The underlying mechanisms of persistence and pathogenesis of infection by HIV and EIAV are similar, albeit the clinical manifestations are different [4]. HIV and EIAV share similar biological properties such as genome structure, viral replication and the conformational structure of some viral structural proteins [10]. Similar to infection by HIV, horses infected by EIAV experience a subclinical phase after recurring cycles of fever, thrombocytopenia, and wasting symptoms [24]. EIAV has also been used as a model to study HIV persistence, pathogenesis and immune response [1, 5].
Recently, a number of studies have shown that the N-linked glycosylation of lentivirus Env associate closely with viral tropism, infectivity and viral escape against the host immune response [12, 13, 15, 19, 23, 27]. Similar to other viruses, tche envelope protein of EIAV plays a critical role in mediating receptor binding to target cells, subsequent entry, and induction of host humoral immune response [3, 16]. The glycosylation modification endows the Env and the viruses a number of biological functions to enhance the binding and infection ability. The potential N-linked glycosylation sites (PNGS) of the EIAV envelope protein play a pivotal role in EIAV attenuated vaccine. This vaccine is highly effective in protecting the vaccinated animals from infection by wild type strains. In comparison with the virulent strain, the attenuated EIAV strain has been shown to have lower numbers of N-linked glycosylation sites [11, 26]. Whether less glycosylation of the EIAV Env contributes to its successes as a vaccine remains to be determined. The effectiveness of the vaccine to protect infection by EIAV virulent strains demonstrated this vaccine can serve as a good model to study the induction of protective immune response against other lentiviruses, including HIV.
Therefore, in this study we attempted to compare the envelope protein of various immunodeficiency viruses such as HIV, SIV and FIV with that of EIAV to gather information about the amino acid length, the number of PNGS and their defining characteristics within Env of these viruses. Furthermore, we analyzed Env sequences to investigate the evolution of these viruses from various lineages. The comparative study of Env from various lineages lentiviruses to EIAV highlighted some common evolutional characteristics that may help to provide some information for the development of an effective HIV vaccine.
HTML
-
HIV-1 env sequences comprising subtypes A-K downloaded from the Los Alamos HIV Database (http://hiv.lanl.gov/content/ index) [9]. In order to improve the accuracy of the phylogenetic analyses, we excluded previously determined recombinant sequences [30]. A total of 48 env from subtype HIV-1 A, B, C, D, F, G, H, J, K, HIV-1 group O and HIV-1 group N were finally selected. Envelope gene sequences of SIV, FIV and EIAV were downloaded from NCBI (http://www.ncbi.nlm.nih.gov).
-
Env sequences were aligned using CLUSTALX [29]. A Neighbor-Joining (NJ) tree was constructed for all the Env sequences which included subtypes A-K of the HIV-1 Env sequences and all of SIV, EIAV and FIV Env sequences by MEGA 4.1 software. We calculated the evolutionary distance through DNAPars DNA parsimony method supplied provided by the BioEdit 7.0.5 software [7, 28].
-
Potential N-linked Glycosylation sites (PNGS) were predicted using N-Glycosite from the Los Alamos National HIV database [32].(http://www. hiv. lanl. gov/content/sequence/ GLYCOSITE/glycosite.html). This is a web-based tool that was developed for tracking and quickly assessing the patterns in N-linked glycosylation sites during protein alignments [32].
-
To compare the amino acid length and the number of PNGS of envelope gene between various immunodeficiency viruses and EIAV, we used a non-parametric Mann-Whitney test. The p values of less than or equal to 0.05 were considered significant. All statistical tests were performed with GraphPad Prism software version 5.0 [31].
Sequences collection
Phylogenetic Analysis
Potential N-linked Glycosylation Sites (PNGS)
Statistical Analysis
-
We analyzed 93 Env sequences in total which included HIV-1 Env (n=48), SIV Env (n=14), FIV Env (n=15) and EIAV Env (n=16, including one EIAV vaccine strain sequence), respectively. The 48 HIV Env came from three groups of M, O and N. They include subtype A, B, C, D, F, G, H, J, K from group M. We built a phylogenetic tree, where each leaf corresponds to a different strain of virus whose trait values are known. We calculated the evolutionary distance reflecting the evolutionary relationship among different strains. As shown in Fig. 1, the evolutionary distance between HIV and SIV was 0.257, followed by the distance between EIAV and FIV, which was 0.52. Slightly higher distance was displayed between FIV and HIV, as well as between FIV and SIV, which was 0.604 and 0.63, respectively. The furthest distance observed was between EIAV and SIV, which was 0.781. A similar distance was observed between EIAV and HIV, which was 0.775.
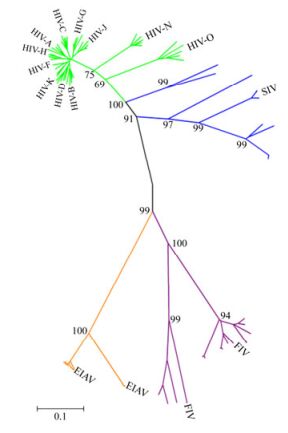
Figure 1. A phylogeny of Env from various lineages of immunodeficiency viruses and EIAV. Each kind of immunodeficiency viruses Env sequences were labeled with different colors (HIV, green; SIV, blue; FIV, purple; EIAV, orange). The black line represents the backbone of the Neighbor-Joining tree. The tree was constructed using MEGA version 4.1.
-
We examined the genetic features of Env including the amino acid length which may influence the protein folding and reflect the diversity of Env.
We found that the amino acids length of EIAV Env ranges from 561 to 865 (median, 709) and was the shortest among the viruses analyzed. The amino acid length of HIV ranges from 812 to 899 (median, 854) was the longest, followed in turns by SIV (ranging from 832 to 897, median, 852), and FIV (ranging from 810 to 863, median, 853). The results showed that the amino acids lengths of SIV and FIV were both shorter than that of HIV, but the difference was not statistically significant (p > 0.05, HIV vs SIV, p > 0.05, HIV vs FIV) (Fig. 2D, 2E). Interestingly, the amino acids length of EIAV Env was significantly shorter than that of HIV, FIV and SIV, respectively. The difference was statistically significant (p < 0.01) (Table 1 and Fig. 2A, 2B, 2C).

Table 1. Comparison of PNGS, length and relative glycosylation density of Env between various immunodeficiency viruses and EIAV
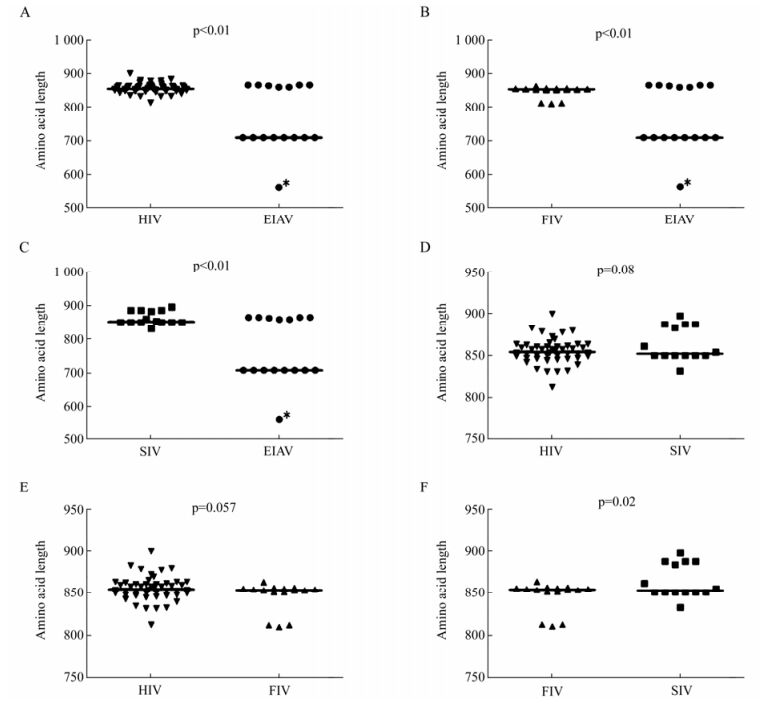
Figure 2. Comparison of Env amino acid length of HIV, SIV, FIV and EIAV. A: Comparison of Env amino acid length between HIV and EIAV. The outlier is labeled with the asterisk (*). B: Comparison of Env amino acid length between FIV and EIAV. C: Comparison of Env amino acid length between SIV and EIAV. D: Comparison of Env amino acid length between SIV and HIV. E: Comparison of Env amino acid length between FIV and HIV. F: Comparison of Env amino acid length between FIV and SIV. The p value for each pair is shown.
-
The glycosylation of Env is also an important factor which affects the function of Env, such as immunogenicity, cell tropism and the replication capacity of the virus. We first compared the consensus Env of SIV and HIV, and we found that the number of PNGS located on the gp120 was more than that on gp41 (Fig. 3). With regard to the number of PNGS, among the different viruses analyzed, the HIV Env exhibited the highest number of PNGS, followed by SIV and FIV. Interestingly, among all the viruses analyzed, EIAV had lowest number of PNGS (p < 0.05) (Table 1 and Fig. 4A, 4B and 4C). With regard to the number of PNGS among the three immunodeficiency viruses (HIV, SIV and FIV), HIV was the most and FIV was the least. There was a reduced number of PNGS in SIV as compared to HIV, which was statistically significant (p < 0.001). Similarly, the number of PNGS in FIV Env was less than that observed in HIV and SIV, and the difference was also significant with HIV (p < 0.001) but not with SIV (p=0.02) (Fig. 4D, 4E and 4F).
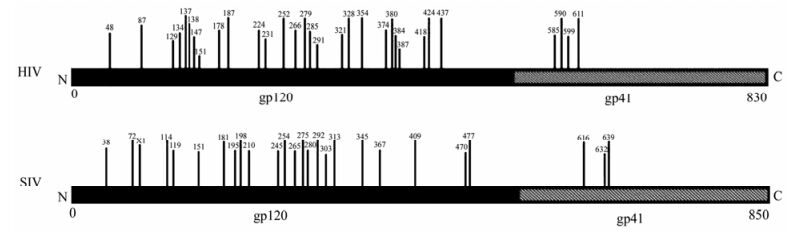
Figure 3. Potential N-linked glycosylation sites distributed in the extracellular membrane gp120 is more than the transmembrane gp41 for HIV and SIV. (Vertical bars denoted potential N-linked glycosylation sites and the numbers above the vertical bars represented the amino acid positions of N-linked glycosylation sites.)
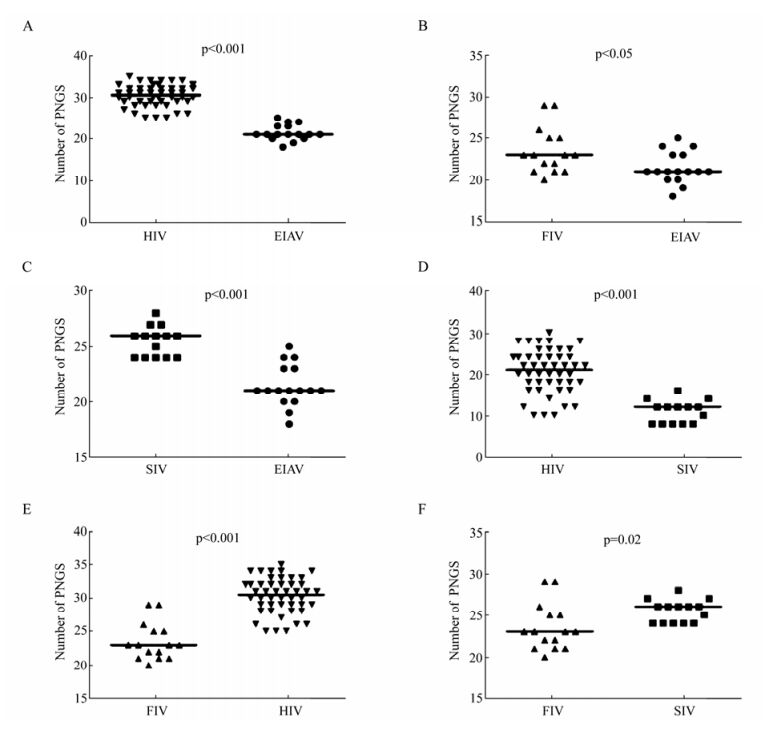
Figure 4. Comparison of PNGS in Env of HIV, SIV, FIV and EIAV. A: Comparison of PNGS in Env between EIAV and HIV. B: Comparison of PNGS in Env between EIAV and FIV. C: Comparison of PNGS in Env between EIAV and SIV. D: Comparison of PNGS in Env between HIV and SIV. E: Comparison of PNGS in Env between HIV and FIV. F: Comparison of PNGS in Env between SIV and FIV. The p value for each pair is shown.
Our results showed that Env quasispecies variety of viruses in Fig. 2. We further compared the relative glycosylation density as normalized to Env amino acids length. Consistent with the total number of glycosylation sites analyzed, the density of HIV glycosylation was the highest among the various lentiviruses analyzed; much higher than those observed with EIAV, with the glycosylation density ranging from 2.10 to 3.39, with SIV or with FIV (p < 0.01) (Table 1 and Fig. 5A, 5D, 5E). A comparison of the other viruses showed that the relative glycosylation density of SIV was slightly higher than that of FIV and EIAV, but there was no significant difference (p > 0.05) (Fig. 5C, 5F).
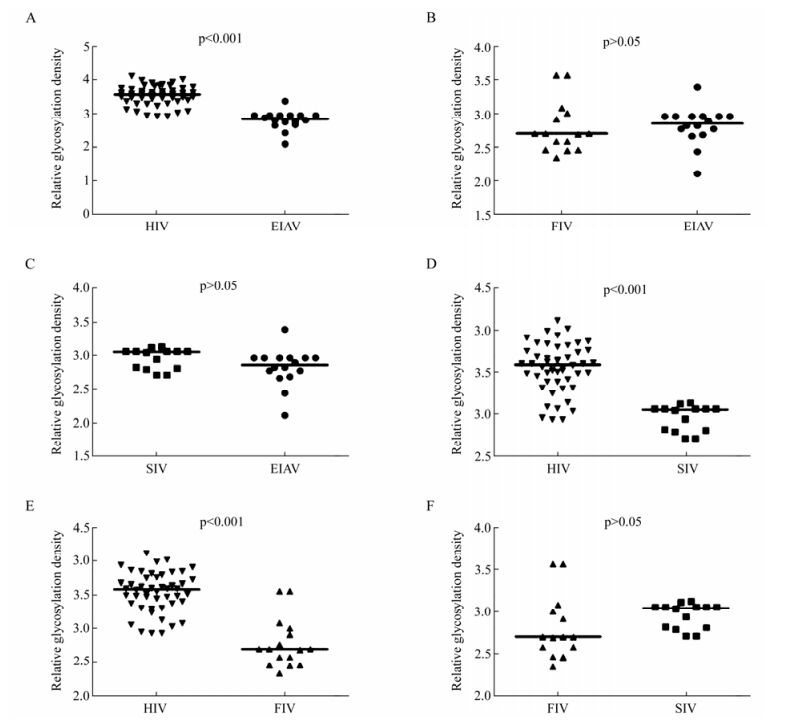
Figure 5. Comparison of relative glycosylation density of potential N-linked glycosylation sites in Env of HIV, SIV, FIV and EIAV. A: Comparison of relative glycosylation density in Env between EIAV and HIV. B: Comparison of relative glycosylation density in Env between EIAV and FIV. C: Comparison of relative glycosylation density in Env between EIAV and SIV. D: Comparison of relative glycosylation density in Env between HIV and SIV. E: Comparison of relative glycosylation density in Env between HIV and FIV. F: Comparison of relative glycosylation density in Env between SIV and FIV. The p value for each pair is shown.
Phylogenetic linkage of Env> between various immunodeficiency viruses and EIAV
Comparison of the amino acids lengths between Env of various immunodeficiency viruses and EIAV
The comparison of the total number and relative glycosylation density of potential N-linked glycosylation sites of Env between various immunodeficiency viruses and EIAV
-
Although the clinical manifestations of infections by immunodeficiency viruses and EIAV are different, the underlying pathogenesis are very similar [4]. The success of EIAV vaccination might facilitate the research on effective HIV vaccine design. As a basis toward defining the role of this protein in the pathogenesis, we studied envelope genotype phylogeny of the lentiviruses, including the various immunodeficiency viruses and EIAV. The phylogenetic traits of those viruses analyses included the env gene evolutionary distance, amino acid length and the number of PNGS.
It is well established that virus evolution is selected for both the host and the virus itself, and Env is more likely to mutate in the variable regions than other viral genes. Therefore, the evolutionary distance of Env will reflect evolutionary relationship among species of viruses. Our results show that the Env protein in HIV was most closely related to SIV and the greatest distance was observed between EIAV and SIV.
The Env of retroviruses have a number of common features. However, the envelope protein of lentiviruses is significantly larger than that of other retroviruses. The size difference suggests that there may be a lentivirus-specific function associated with Env [22]. Among various immunodeficiency viruses and EIAV analyzed, the Env length of EIAV was the shortest and HIV was the longest. It suggests that the immunodeficiency viruses are more complex than EIAV with respect to the Env amino acid length, and it is possible that such complexity of the Env amino acid sequence length could lead to more sequence variation and diversification for the HIV Env, which is a major obstacle in developing an effective HIV vaccine.
The N-linked glycosylation sites influence the envelope protein to fold correctly and exert the normal functions. More important, the deglycosylation of the envelope proteins is closely related to an attenuated vaccine [18]. Our study has demonstrated that the number of PNGS in immunodeficiency viruses of primates (human and simian) is more than that of feline and equine. Because of the Env protein exists as quasispecies, the comparison of PNGS was normalized to Env amino acid length in order to compare the density of glycosylation. The relative glycosylation density of HIV and SIV was higher than that of FIV and EIAV, especially for HIV. We found that the difference of PNGS between HIV and SIV was mainly on gp120. There have been reports that the surface glycoprotein for the virulence was stronger than transmembrane glycoprotein [20]. Previous studies have shown that the heavy glycosylation of Env is an unique feature of HIV/SIV that is distinct from other enveloped viruses, and it is significantly related to their neutralization resistant properties [2, 14, 21]. This study showed that the number of PNGS and the density of glycosylation is also the lowest after normalization with the Env sequence length. Therefore, we speculate that the effective EIAV attenuated vaccine benefits from the less number of PNGS in Env and the less relative glycosylation density. In agreement with, the insufficient immunosuppression of HIV/SIV might be due to heavy glycosylation sites in Env and remove some of the glycans could enable the host to mount the stronger protective immune response against the infection [18]. Currently, the heavily glycosylated structure of Env has been considered as a main contributor to chronic and persistent viral replication, and the pathogenicity of HIV or SIV, primarily because it potentially interferes with the development of an effective host immune response [6, 17, 18].
Our results showed that there was a significant difference in the number of PNGS between HIV and SIV in spite of similar amino acid length of Env. The number of PNGS in SIV was significantly less than that of HIV. The failure of HIV vaccine in clinical trial might be explained by the difference of PNGS in Env between simian and human. The quasispecies of Env as a result of amino acid length and the number of PNGS variable are an obstacle for successful HIV/AIDS vaccine. It is possible that the successful EIAV Env attenuated vaccine strains possess an advantage over other immunodeficiency viruses because EIAV Env sequences have shorter amino acid length and less number of potential N-linked glycosylation sites (PNGS). The modification of glycosylation of HIV vaccine might be different between human and simian, even though they had been tested effective in simian. In addition, the glycosylation sites are known for play an important role in viral sensitivity to neutralization, pathogenicity and the cell tropism [8].













 DownLoad:
DownLoad: