HTML
-
We report the results of cloning, expression, subcellular localization analysis, and molecular properties of the Epstein-Barr virus (EBV) BFRF3 gene, using several bioinformatics tools. Bioinformatics analysis indicated that the encoding protein of EBV BFRF3 gene (designated BFRF3) has a conserved Herpes_capsid domain, which was found to be closely related to the gammaherpesvirus capsid protein family, and is highly conserved among its counterparts. Multiple nucleic acid sequence and amino acid (aa) sequence alignments suggested that BFRF3 has a relatively higher homology with BFRF3-like proteins of the genus Lymphocryptovirus than with that of other genera of Gammaherpesvirinae. In addition, phylogenetic analysis showed that BFRF3 has a close evolutionary relationship withmembers of Gammaherpesvirinae, especially members of the genus Lymphocryptovirus of Macacine herpesvirus 4. Antigen prediction demonstrated that there are several potential B-cell epitopes located in BFRF3. Furthermore, secondary structure prediction revealed that BFRF3 consists predominantly of α-helices.
EBV, the causative agent of infectious mononucleosis, has a global distribution in the human population as a life-long and largely asymptomatic infection. EBV is reported to be associated with various malignancies, including epithelial tumor, nasopharyngeal carcinoma, and a subset of T-cell and natural killer cell lymphomas. It may also be related to three B-cell associated malignancies: Burkitt's lymphoma, Hodgkin's lymphoma, and post-transplant lymphoproliferative disease(Henderson S, et al., 1991; Iwakiri D, et al., 2005; Long H M, et al., 2011; Niller H H, et al., 2011; Ruf I K, et al., 2000; Samanta M, et al., 2006; Shimizu N, et al., 1994).
EBV assembles icosahedral capsid structures in the nuclei of infected cells. Six proteins are required for the assembly of these structures, including BcLF1, BORF1, BDLF1, BdRF1, BVRF2, and BFRF3. Although the small capsid protein BFRF3 is required for the stable capsid assembly structure (Henson B W, et al., 2009), the specific functions of the BFRF3 gene and its protein product BFRF3 are less understood.
To obtain the BFRF3 gene, we performed PCR using purified BAC DNA of the EBV Akata strain as the template and primers BFRF3-F (5′-TTGAATTCCATGGCACGCCGGCTGCCCAAG) and BFRF3-R (5′-AAGGATCCCTACTGTTTCTTACGTGCCCC), where the restriction enzyme sites EcoR I and BamH I are underlined. The BFRF3 fragment (531 bp) was then cloned into pEYFP-C1 (Clontech, Mountain View, CA, USA) to yield pEYFP-BFRF3, which was verified by restriction digestion analysis, PCRamplification, and DNA sequencing.After sequence confirmation, the recombinant plasmid pEYFP-BFRF3 was transfected into HEK293T cells, and western blotting using a monoclonal antibody against YFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA) as described prev iously(Li M L, et al., 2011; Li M L, et al., 2011), confirmed that the open reading frame of BFRF3 was properly expressed in the transfected HEK293T cells (approximately 48 kDa). In addition, subcellular localization analysis showed that EYFP-BFRF3 was distributed mainly in the nucleus, as previously described (Henson B W, et al., 2009).
Nucleotide sequence similarity search using NCBI BLASTN yielded five nucleotide sequences with strong similarity to the EBV Akata strain BFRF3 (98.9%, 99.1%, 99.2%, 99.4%, and 100%, respectively); these sequences corresponded to BFRF3 of the EBV B95-8, AG876, Mutu, HKNPC1, and GD1 strains respectively. Multiple alignments of EBV BFRF3 with 10 homologous reference gammaherpesviruses revealed a remarkably high homology of 70% with members of the genus Lymphocryptovirus, subfamily Gammaherpesvirinae, that is, Macacine herpesvirus 4 (McHV-4). However, relatively low homology was detected between EBV and other members of the subfamily Gammaherpesvirinae.
An aa sequence similarity search using NCBI BLASTP yielded five aa sequences that strongly matched the target sequence of EBV Akata BFRF3(99.4%, 100%, 100%, 100%, and 100%, respectively), and these correspondedto the BFRF3 protein of the EBV B95-8, AG876, Mutu, HKNPC1, and GD1 strains, respectively. these results clearly demonstrated that the Akata strain is the most closely related strain to GD1, which is consistent with a previous report (Lin Z, et al., 2013); the results also revealed close relationships among the studied EBV strains. In addition, multiple sequence alignments of BFRF3 with its homologs in 10 reference gammaherpesviruses showed relatively high homology of 38.6 to 70% between BFRF3 and its Callitrichine herpesvirus 3(CalHV-3) and McHV-4 counterparts. However, BFRF3 shared no substantial homology with BFRF3-like proteins from Ovine herpesvirus 2 (OvHV-2) or HHV-8, with values of only 19.9% and 22.9%, respectively. Therefore, BFRF3 has high homology with Lymphocryptovirus but not with Rhadinovirus or Macavirus.
Analysis with Clustal X (version 1.8) was used to compare multiple sequence alignments of the EBV Akata strain BFRF3 with its 15 homologs and the result demonstrated that the most conserved region was the N-terminus of 16 BFRF3-like proteins (Fig. 1A). In agreement with this, conserved domain analysis indicated that BFRF3 contained an apparent conserved domain of Herpes_capsid (Fig. 1B), which is a member of the gammaherpesvirus capsid protein family and is locatedin the N-terminal of BFRF3. Accordingly, BFRF3 may belong to the Herpes_capsid superfamily, and the N-terminus of BFRF3 might be important for achieving its functions.
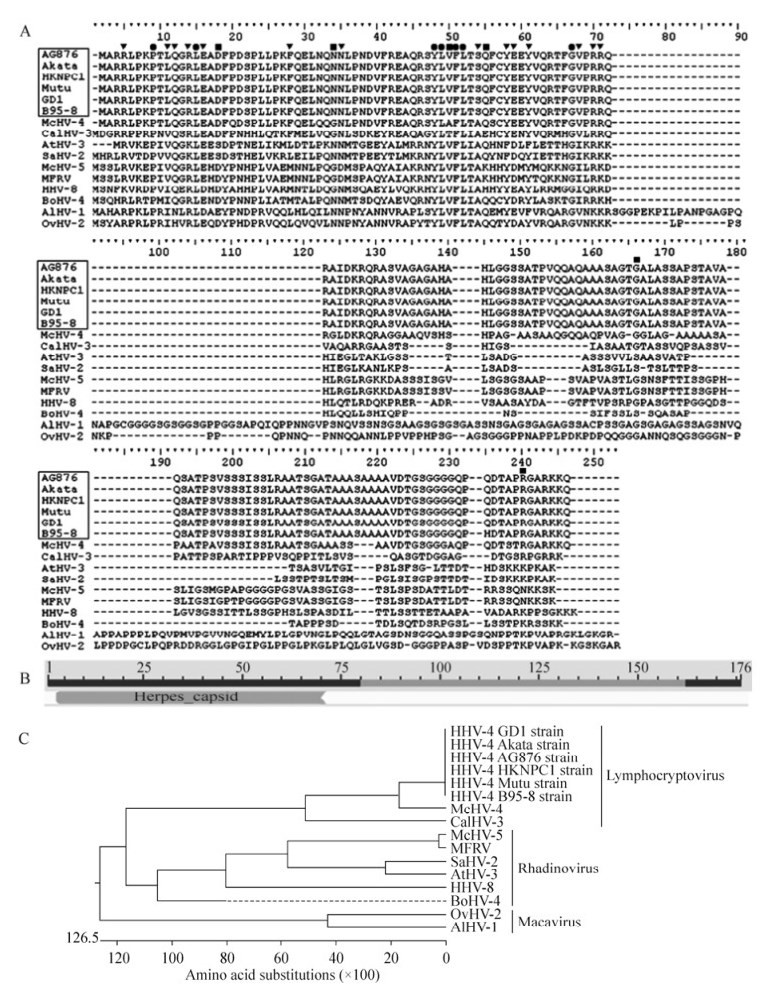
Figure 1. Multiple amino acid (aa) sequence alignments, conserved domain analysis, and phylogenetic relationships of Epstein-Bar virus (EBV) BFRF3 with other BFRF3-like proteins from 15 reference species. A. Multiple aa sequence alignments of EBV BFRF3 with its homologous proteins from 15 selected species (Table 1) using Clustal X (version 1.8) and Bioedit 7.0. Gaps were introduced by the alignment program to maximize the homology. "●" indicates positions that have a single, fully conserved residue between the 16 BFRF3-like proteins. "▼" indicates conservation of strong groups, "■" indicates conservation of weak groups. Viruses with a box indicate different strains from the same virus EBV. B. Conserved domain analysis of EBV BFRF3 using the NCBI Conserved Domains search tool. C. Phylogenetic relationships of the putative EBV BFRF3 protein with other BFRF3-like proteins from 15 Gammaherpesviruses (Table 1). The phylogenetic tree of these proteins was generated using the Megalign program in LASERGENE (DNAStar version 7.0) with the Clustal V method, and sequence distance was calculated using the structural weight matrix. Gaps were introduced by the alignment program to maximize the homology.
Sub-family Genus Virus name (Abbreviation) Strain Natural host GenBank accession number Gammaherpesvirinae Lymphocryptovirus Human herpesvirus 4 (HHV-4); Epstein-Barr virus (EBV) Akata Homo sapiens (Human) AFY97839 GD1 AAY41101 HKNPC1 AFJ06843 Mutu AFY97924 AG876 ABB89228 B95-8 CAD53401 Lymphocryptovirus Macacine herpesvirus 4 (McHV-4); Rhesus lymphocryptovirus (LCV) LCL8664 Macaca mulatta (Rhesus monkey) YP_067949 Lymphocryptovirus Callitrichine herpesvirus 3 (CalHV-3); marmoset lymphocryptovirus (MLCV) CJ0149 Callithrix jacchus (Common marmoset) NP_733913 Rhadinovirus Bovine herpesvirus 4 (BoHV-4) V.test Bos taurus (Domestic cow) AEL29809 Rhadinovirus Macacine herpesvirus 5 (McHV-5); Rhesus rhadinovirus (RRV) 17577 M. mulatta NP_570813 Rhadinovirus Macaca fuscata rhadinovirus (MFRV) Macaca fuscata (Japanese monkey) AAT00110 Rhadinovirus Saimiriine herpesvirus 2 (SaHV-2); herpesvirus saimiri (HVS) 11 Saimiri sciureus (Squirrel monkey) NP_040267 Rhadinovirus Human herpesvirus 8 (HHV-8); Kaposi's sarcoma-associated herpesvirus (KSHV) GK18 H. sapiens YP_001129422 Rhadinovirus Ateline herpesvirus 3 (AtHV-3); herpesvirus ateles (HVA) 73 Ateles spp, (Spider monkey) NP_048036 Macavirus Alcelaphine herpesvirus 1 (AlHV-1); malignant catarrhal fever virus (MCFV) C500 Connochaetes taurinus taurinus (Wildebeest) NP_065564 Macavirus Ovine herpesvirus 2 (OvHV-2) Ovis aries (Sheep) ABB22284 Table 1. Abbreviations and GenBank accession numbers for 16 BFRF3-like proteins from different species
Phylogenetic analyses of EBV and other gammaher-pesviruses were performed based on the sequences of BFRF3 and the BFRF3-like proteins of 15 reference gammaherpesviruses (Table 1). The proteins were preliminarily separated into different genera, namely Lymphocryptovirus, Rhadinovirus, and Macavirus, consistent with the existing classification of the subfamily Gammaherpesvirinae (Fig. 1C). Furthermore, the EBV Akata, B95-8, AG876, Mutu, HKNPC1, and GD1 strains were found to be different from other gammaherpesviruses. They clustered together, forming a separate branch, and also clustered with members of the genus Lymphocryptovirus, such as McHV-4 and CalHV-3. Subsequently, they clustered with other members of the genera Rhadinovirus and Macavirus (Fig. 1C). Therefore, EBV might have a closer evolutionary relationship to members of genus Lymphocryptovirus than to members of other genera of Gammaherpesvirinae.
Signal polypeptide prediction indicated no signal polypeptide cleavage site or potential transmembrane domain in BFRF3. Furthermore, N-linked glycosylationsite (Asn-X-Ser/Thr) prediction did not identify an N-glycosylation site in BFRF3, confirming the absence of a signal peptide in BFRF3, and BFRF3 may be a real capsid protein but not a glycoprotein (Henson B W, et al., 2009). Interestingly, nine potential phosphorylation sites were found in BFRF3: one tyrosine, one threonine, and seven serine residues. It is well known that tyrosine phosphorylation is involved in the modification of protein translocation from the cytoplasm to the nucleus during the course of productive viral infection, as well as during the replication of several herpesviruses. It has been reported that phosphorylation occurs on both the serine and threonine residues of HSV-1 UL35, which are associated with the morphology of HSV-1. Thus, BFRF3 phosphorylation may also play an important role during EBV infection, perhaps by modulating its proper localization, stable capsid structure assembly, or other uncharacterized functions, such as a potential regulatory role in the affinity of BFRF3 for DNA (McNabb D S, et al., 1992).
Hydrophobicity analysis revealed threehydrophobic regions located at aa 45 to 56, 84 to 91, and 158 to 106. Compared with the hydrophobic region, the proportionality of the hydrophilic region was nearly in conformity with the hydrophobic region, which might represent the internal and surface regions of the protein. In addition, analysis of a potential B-cell epitope determinant demonstrated several potential B-cell epitopes in BFRF3, situated in or adjacent to aa 0 to 24, 26 to 48, 65 to 83, 139 to 146, and 155 to 176, which might yield methods for developing new antibodies and immunoassays for application in the clinical diagnosis of EBV. Secondary structure analysis suggested that BFRF3 primarily consisted of α-helices (59.1%) and random coils (36.9%), whereas a b-strand accounted for the least prevalent component (4.0%). The 3D structure prediction revealed no 3D structure model for BFRF3. Therefore, the principal component of the α-helix ofBFRF3, which contains many highly conserved residues, might be engaged in its self-association or in interactions with other nucleocapsid components(Benach J, et al., 2007; Ohta A, et al., 2011), tegument or host proteins (Uesugi M, et al., 1997).
In conclusion, we present the results of cloning, expression, and molecular characterization of the EBV BFRF3 gene. Elucidating the relationship between the molecular characterization and genetic evolution of EBV BFRF3 will provide insights for further research on the function and mechanism of BFRF3 during EBV infection. Such studies will contribute to our understanding of this virus at the molecular level, as well as enrich the herpesvirus database.
-
This work was supported by grants from Science and Technology New Star in Zhu Jiang, Guangzhou City (2013J2200018); the Natural Science Foundation of Guangdong Province (S2013040016596); the National Natural Science Foundation of China (31200120); the Medical Scientific Research Foundation of Guangdong Province, China (B2012165); the Foundation for Distinguished Young Talents in Higher Education of Guangdong, China; First Batch of Young Core Instructor of Guangzhou Medical University; the Science and Technology Foundation for Universities in Guangzhou, China (10A173 and 08A097); and the Students' Extracurricular Scientific And Technological Activities in Guangzhou Medical University (2012A039 and 2012C007). We thank Dr. Teru Kanda for the generous gift of Akata-derived EBV-BAC.
-
Mingsheng Cai, Zhiyao Zhao, and Wei Cui carried out most of the experiments. Mingsheng Cai and Meili Li wrote the manuscript and have critically revised the manuscript and the experimental design. Lin Yang, Junyi Zhu, Yalan Chen, Changling Ma, and Zhuqing Yuan helped in experiments. All the authors read and approved the final manuscript.













 DownLoad:
DownLoad: