HTML
-
Hepatitis C virus (HCV), which is an enveloped, positive-strand RNA virus that belongs to the Flaviviridae, is a major cause of hepatitis that frequently progresses to cirrhosis and to hepatocellular carcinoma (Alter H J, et al., 2000; Shimotohno K, 2000). The RNA genome of HCV is approximately 10 kb in length and consists of a single open reading frame that encodes a polyprotein of approximately 3, 000 amino acids (aa), which is then processed into viral structural and non-structural proteins by host and viral proteinases (Grakoui A, et al., 1993; Hijikata M, et al., 1993).
The replicase of HCV is composed of some, if not all, of the viral nonstructural (NS) proteins, and possibly some host factors also (Lohmann V, et al., 1999). Amongst these, two of the viral proteins that possess enzyme activity, NS3 and NS5B, play a key role. The N-terminal region (approx. one third) of NS3 functions as a serine proteinase, together with the co-factor NS4A, whereas the C-terminal (approx. two-thirds) exhibits RNA helicase and RNA-stimulated NTPase activities. The NS5B protein exhibits an RNA-dependent RNA polymerase activity (Lindenbach B D, et al., 2005). The proteinase and helicase domains of NS3 are reported to enhance the activity of each other (Beran R K, et al., 2008; Beran R K, et al., 2007); the proteinase domain alone is reported to bind HCV RNA and to regulate replication and translation (Ray U, et al., 2011).
Since the first construction of an HCV subgenomic replicon in 1999 (Lohmann V, et al., 1999) and the first establishment of an infective HCV cell-culture system in 2005 (Wakita T, et al., 2005), many chimeric and adaptive mutated constructs have been reported to replicate in, or to infect, a variety of cell lines (Gottwein J M, et al., 2007; Gottwein J M, et al., 2009; Murayama A, et al., 2012; Pietschmann T, et al., 2006; Scheel T K, et al., 2008; Scheel T K, et al., 2011). In addition, some strains of genotypes 1a, 1b, 2a and 2b that were previously incapable of replication in vitro have been reported to be adapted to cell lines (Date T, et al., 2012; Li Y P, et al., 2012; Pietschmann T, et al., 2009; Ramirez S, et al., 2013; Yi M, et al., 2006).
In this study, an HCV strain denoted as strain CCH was isolated from a Chinese patient with chronic hepatitis C. Various chimeric replicons of JFH1 (AB047639) and CCH were constructed, and several mutations were introduced in order to generate a structure possessing the ability to replicate.
-
Serum was collected in 2008 from a female with chronic hepatitis C. The HCV RNA copy number was 1.39×107 copies/mL and serum levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were normal. Hepatitis B virus infection was discounted, since HBsAg, HBsAb, HBeAg, HBeAb and HBcAb were all negative in the serum.
-
Huh7.5.1 cells, kindly supplied by Prof. Frank Chisari (Zhong J, et al., 2005), were cultured in Dulbecco's modified Eagle medium (DMEM) (GE healthcare), supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/mL of penicillin and 100 U/mL of streptomycin, at 37 ℃ in a 5% (v/v) CO2 atmosphere.
-
Viral RNA was recovered from the serum of the patient using a High Pure Viral RNA Kit (Roche). It was then reverse-transcribed with M-MLV (Promega) using random primers, cloned in fragments using primers based on JFH1 and genotyped by sequencing. After sequencing, the viral RNA was reverse-transcribed, using SuperScriptTM Ⅲ (Invitrogen) with the specific primer 9470R (Table 1). Using nested PCR with LA Taq (Takara) and the primer pairs: outer, S17 + R5239, and inner, S23 + R5285; outer, S4675 + R9440, and inner, S4741 + R9398 (Table 1), two fragments were generated and cloned into pGEMTM-T Easy (Promega). Five clones of each fragment were then sequenced in order to generate a consensus sequence.
Primer Sequence 9470R 5' CTATGGAGTGTACCTAGTGTGTGC 3' S17 5' GGCGACACTCCGCCATGAATC 3' R5285 5' TGTACTTCGTCCCAGGGTGTGTGAG 3' S23 5' ACTCCGCCATGAATCACTCCCCT 3' R5239 5' GCCCAAACGGTACAGGAGAGGTG 3' S4675 5' GACGGGGTACACTGGAGACTTTGA 3' R9440 5' ACCGAGCGGGGAGTAGGAAGAGG 3' S4741 5' CTTCAGCCTGGACCCCACCTTC 3' R9398 5' GGAGTAGGCCGAAGAGTAATGAGC 3' EI/AvrII-S 5' ATTCCTAGGCCTCTCCCTCCCCCCCCCCTAACGTT 3' EI/NS3-R 5' GGGGGCCATGGTATCATCGTGTTTTTCAAA 3' EI/NS3-S 5' CACGATGATACCATGGCCCCCATCACTGCT 3' NS3/KpnI-R 5' GGCGGTACCCAACCTGGTAGGTCTGGGGC 3' Luc-S 5' AACCGTCGCCCACCATGGAAGACGCCAAAAACATA 3' Luc-A 5' GCCGTTTAAACTTACACGGCGATCTTTCCGCCCTT 3' J/C7680-s 5' CCTGGACCGGGGCTCTAATAACTC 3' J/C7703-a 5' GAGTTATTAGAGCCCCGGTCCAGG 3' J/C6248-s 5' GAGGACTGCCCCATCCCATGC 3' J/C6268-a 5' GCATGGGATGGGGCAGTCCTC 3' J/C5325-s 5' GCACGTGGGTCCTAGCTGGAGGAGTC 3' J/C5350-a 5' GACTCCTCCAGCTAGGACCCACGTGC 3' J/C5459-s 5' GAGGCTTTTGATGAGATGGAGGAATG 3' J/C5484-a 5' CATTCCTCCATCTCATCAAAAGCCTC 3' J/C4127-s 5' GCTGCCACCCTGGGGTTTGGGGCG 3' J/C4150-a 5' CGCCCCAAACCCCAGGGTGGCAGC 3' J/C5025-s 5' GGGAGGCAGTTTTCACCGGCC 3' J/C5045-a 5' GGCCGGTGAAAACTGCCTCCC 3' J/C8971-s 5' CCAAGACACCCTGGACCAGAACC 3' J/C8993-a 5' GGTTCTGGTCCAGGGTGTCTTGG 3' CN3-P29Q-s 5' GCGACAAGACCGAACAGGCCGGGGAAATCC 3' CN3-P29Q-a 5' GGATTTCCCCGGCCTGTTCGGTCTTGTCGC 3' CN3-P47T-s 5' CTTTCCTCGGAACGACTATTTCGGGGG 3' CN3-P47T-a 5' CCCCCGAAATAGTCGTTCCGAGGAAAG 3' CN3-D71V-s 5' CACGGGGTCCGGTCACACAGATGTACTC 3' CN3-D71V-a 5' GAGTACATCTGTGTGACCGGACCCCGTG 3' CN3-N185S-s 5' GGTCTCCCACCTTCAGTGACAACAGCTCACC 3' CN3-N185S-a 5' GGTGAGCTGTTGTCACTGAAGGTGGGAGACC 3' CN3-R246H-s 5' CTTGTCCAAGGCACATGGCATCAACCC 3' CN3-R246H-a 5' GGGTTGATGCCATGTGCCTTGGACAAG 3' CN3-D281S-s 5' GATGGGGGCTGTGCAAGCGGCGCCTATGACG 3' CN3-D281S-a 5' CGTCATAGGCGCCGCTTGCACAGCCCCCATC 3' CN3-T347I-s 5' CAGGAGGGCGAGATCCCCTTCTATGGG 3' CN3-T347I-a 5' CCCATAGAAGGGGATCTCGCCCTCCTG 3' CN3-S497A-s 5' GTGAGTGCTACGACGCAGGGGCTGCATGG 3' CN3-S497A-a 5' CCATGCAGCCCCTGCGTCGTAGCACTCAC 3' C5B-T35N-s 5' GCTGTTGCGATACCACAACAAGGTGTACTGTACTAC 3' C5B-T35N-a 5' GTAGTACAGTACACCTTGTTGTGGTATCGCAACAGC 3' C5B-A179T-s 5' GGCCCTTTATGATGTCACACAAAAGCTCCCTC 3' C5B-A179T-a 5' GAGGGAGCTTTTGTGTGACATCATAAAGGGCC 3' C5B-A322V-s 5' GCGACGACTTGGTTGTCATCTCAGAGAGCC 3' C5B-A322V-a 5' GGCTCTCTGAGATGACAACCAAGTCGTCGC 3' C5B-E310A-s 5' GCAGGGATAGTTGCGCCCACGATGCTG 3' C5B-E310A-a 5' CAGCATCGTGGGCGCAACTATCCCTGC 3' C5B-V405I-s 5' CTAGACACTCCCCTATCAATTCATGGCTGG 3' C5B-V405I-a 5' CCAGCCATGAATTGATAGGGGAGTGTCTAG 3' J5166-s 5' ACGCCATGTGGAAGTGCCTGGCCCG 3' C4452-a 5' AGGGCTACCTCCTCTATATTGGGG 3' C4220-s 5' CCCATCACATACTCCACATATGGC 3' C8486-a 5' AGGTCTGACCCTTGCTGTTAAACATGG 3' C8477-s 5' GGTCAGACCTGCGGGTACAGGCGTTG 3' M13-R 5' CAGGAAACAGCTATGACC 3' Table 1. Primers used in this study
-
The full-length JFH1 plasmid, pJFH1, was kindly supplied by Prof. Wakita Takaji (Wakita T, et al., 2005). The primers used for the construction of the CCH cDNA clone, for chimeric replicon construction and for site-directed mutagenesis are listed in Table 1. The subgenomic HCV replicons, pSGR-Luc-JFH1 and pSGR-Luc-CCH, were constructed according to a previous method (Kato T, et al., 2003), using the primers EI/AvrII-S, EI/NS3-R, EI/NS3-S, NS3/KpnI-R, Luc-S and Luc-A (Table 1). For the construction of chimeric replicons, primers for recombinant PCR were designed within the regions of complete identity between JFH1 and CCH, as indicated in Table 1. The different recombinant fragments were then cloned into the unique restriction-enzyme sites of pSGR-Luc-JFH1 in order to generate the chimeric replicons. Specific mutations were introduced by the use of the corresponding site-directed mutation primers listed in Table 1, followed by cloning as above.
-
The plasmids were linearized and transcribed in vitro as described previously (Han Q, et al., 2009). For RNA electroporation, trypsinized Huh7.5.1 cells were washed twice with PBS and then resuspended in Cytomix (120 mM KCl; 0.15 mM CaCl2; 10 mM K2HPO4/KH2PO4 (pH 7.6); 25 mM Hepes; 2 mM EGTA; 5 mM MgCl2) at a density of 1.0×107 cells/mL. Cell suspension (400 μL) was then mixed with in vitro-transcribed HCV genomic RNA (10 μg) and pulsed (270 V, 975 μF) using a Gene Pulser Xcell™ apparatus (Bio-Rad). The electroporated cells were then immediately transferred into complete DMEM. For the Luciferase assay, cells were seeded onto 24-well plates and luciferase activity was measured at specified time points post-electroporation, using a Steady-GloTM luciferase assay system (Promega). For colony formation assay, electroporated cells were seeded into 10 cm dishes and selected with medium containing 1000 IU/L of G418 Sulfate (G418) (Inalco) 2 days after electroporation for 3 weeks. The cells were rinsed with PBS, fixed with methanol, and stained with 0.5 % (w/v) crystal violet.
Serum
Cell culture
cDNA cloning
Replicon construction
RNA transcription and electroporation
-
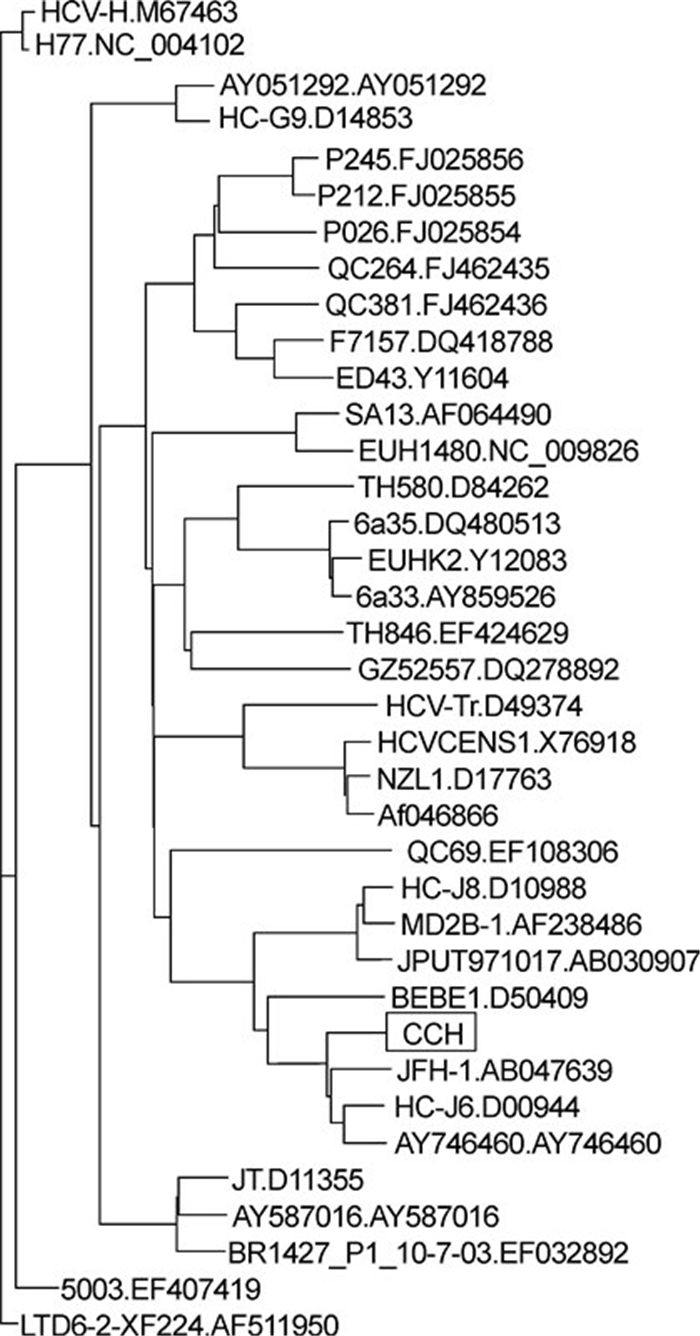
The HCV in serum obtained from a chronic hepatitis C patient with a high serum-HCV copy number ( > 107/mL) was identified as genotype 2a, by sequence alignment of the core regions amplified using conserved primers (data not shown). The complete open reading frame, encoding 3033 aa and partial untranslated regions (UTRs) of the HCV, was then amplified using genotype 2a-based primers. The consensus sequence was constructed based on the sequences of five clones. To elucidate the relationship between the consensus sequence and other reference sequences, a phylogenetic tree was constructed (Figure 1) using the Neighbor TreeMaker tool of the HCV Database (Kuiken C, et al., 2005). This revealed that the strain, denoted as CCH (from Chinese Chronic Hepatitis), was clustered into genotype 2a and showed a nucleotide sequence identity of 89% with strain JFH1 (AB047639) and of 92% with strain J6CF (D00944).
-
In view of its high viral titer in serum and its high degree of similarity to JFH1, it was anticipated that CCH might replicate efficiently in vitro. Therefore, in order to determine the replication capability of the CCH strain, a subgenomic replicon construct containing the minimal viral replicase NS3 to NS5B, of HCV and a firefly luciferase reporter gene (pSGR-Luc-CCH) was constructed (Figure 2A). The replication of SGR-luc-CCH was determined by monitoring the luciferase level in Huh7.5.1 cells electroporated with in vitro-transcribed RNA (Figure 2B). The constructs pSGR-luc-JFH1 and pSGR-luc-JFH1GND (A point mutation in NS5B from GDD to GND, which abolish the polymerase activity) were used as positive and negative controls, respectively. As shown in Figure 2B, in SGR-luc-JFH1 RNA-transfected cells, the luciferase activity dropped a little at 24 h post-electroporation and then rose to a high level, whereas in both the SGR-luc-CCH RNA-and SGR-luc-JFH1GND RNA-transfected cells, luciferase activity declined continuously after electroporation, indicating that there was no replication for a period of 96 h post-electroporation. Thus, the SGR-luc-CCH was therefore demonstrated to be replication-defective in Huh7.5.1.
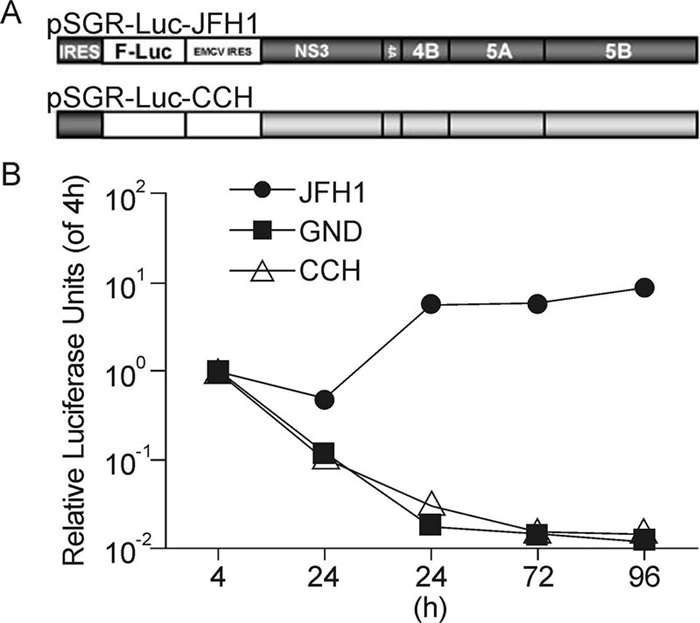
Figure 2. Replication activity of the CCH subgenomic replicon. (A) Schematic structures of the JFH1 and CCH subgenomic replicons. (B) Replication activity of the CCH subgenomic replicon. Huh7.5.1 cells were electroporated with the in vitro-transcribed replicon RNAs as shown and the luciferase activity was monitored in each case at the time points indicated. The results are expressed as luciferase activity, relative to the 4 h value.
-
To define the regions responsible for the defective replication of the CCH replicon, the NS3, NS4A-4B, NS5A, NS5B and NS4A to NS5A regions of pSGR-luc-JFH1 were replaced with the corresponding sequences of CCH (Figure 3A). The replication of these constructs was then determined by luciferase assay. Substitution of either NS3 or NS5B completely abolished the replication of JFH1, while substitute of NS4A to NS4B, NS5A and NS4A to NS5A presented a similar replication level with JFH1 (Figure 3B). This result indicated that the defect of CCH in replication was probably caused by NS3 and/or NS5B; both are key proteins of the replicase. To pinpoint further the sequences responsible, chimeric replicons with different fragment substitutions in NS3 and NS5B were constructed, denoted as JFH1/CN3-1, JFH1/CN3-2, JFH1/CN3-3, JFH1/CN5B-1 and JFH1/CN5B-2, respectively (Figure 3A). Replacing the sequences aa 1031~1262 (JFH1/CN3-1), aa 1263~1561 (JFH1/CN3-2) (in NS3) or aa 2443~2877 (JFH1/CN5B-1) (in NS5B) but not the aa 1563~1661 (JFH1/CN3-3) (in NS3) and aa 2878~3013 (JFH1/CN5B-2) (in NS5B) of SGR-luc-JFH1 with the corresponding CCH sequences completely abolished the replication (Figure 3C and D). Thus, the defective regions of CCH were proven to lie within the sequences aa 1031~1262 and 1263~1561 in NS3, and aa 2443~2877 in NS5B.
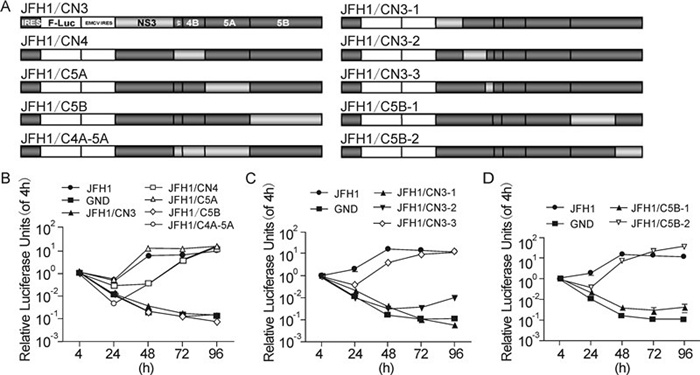
Figure 3. NS3 and NS5B are responsible for the replication deficiency of CCH. (A) Schematic structures of the JFH1 and CCH constructs and of the chimeric replicons. (B) – (D) Replication activities of the chimeric replicons. The results are expressed as luciferase activity, relative to the 4 h value in each case.
-
To predict the amino acids that were potentially responsible for the replication deficiency, the NS3 and NS5B sequences of CCH were used to search against 200 reference sequences in the HCV Database (Kuiken C, et al., 2005). Twelve amino acid residues in CCH that were different with all 200 reference sequences were selected and marked as "CCH Unique Sites"; and these residues were therefore earmarked as potentially responsible for the replication deficiency (Table 2). Residues at the CCH unique sites of JFH1/CN3-1, JFH1/CN3-2 and JFH1/CN5B-1 were then mutated into the corresponding amino acids of JFH1 in different combinations, by sitedirected mutagenesis, as indicated in Figure 4, and the replication capability of each of the replicons was analyzed. For the construct pSGR-luc-JFH1/CN3-1, in which there had been a partial substitution with CCH NS3 (aa 1031~1262), the introduction of the mutations P1059Q and P1077T (pSGR-luc-JFH1/CN3-1 QT) effectively rescued replication from a low level; the introduction of an additional D1101V mutation (pSGR-luc-JFH1/ CN3-1 QTV) then further enhanced the replication level of the chimeric replicon to a level that was comparable to that of JFH1 (Figure 4A). For the construct pSGR-luc-JFH1/CN3-2, in which aa 1263~1561 of CCH NS3 had been substituted, the introduction of R1276H and D1311S (pSGR-luc-JFH1/CN3-2 HS) restored luciferase expression to the same level as that of pSGR-luc-JFH1 at 96 h post-electroporation, although a remarkably lower value was observed between 24 h and 72 h post-electroporation (Figure 4B). For the replicon carrying the substitution aa 2443~2877 of CCH NS5B, a significant increase in luciferase activity was found when the three mutations A2621T, A2764V and T2477N were introduced (Figure 4C). So far, therefore, all three replication-defective chimeric replicons that incorporated fragment substitutions could be rescued by modifying their CCH unique sites.
Amino acid residue (JFH1) CCH JFH1 Con1 H77 NS3 1059 P Q V V 1077 P T C C 1101 D V I V 1215 N S T T 1276 R H H H 1311 D S G G 1377 T I I I 1527 S A A A NS5B 2477 T N N N 2621 A T V V 2752 E A D D 2764 A V V V The CCH NS3 and NS5B sequences were searched against the HCV Sequence Database, and a number of CCH unique sites that differed from all of the reference sequences were revealed. Amino acid residue numbers refer to the JFH1 reference sequence (AB047639). Table 2. Sequence analysis of CCH NS3 and NS5B
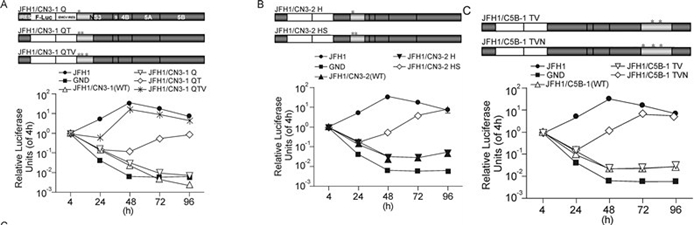
Figure 4. Replication activity of chimeric replicons containing introduced mutations. (A-C: Upper) Schematic structures of the chimeric replicons and of the introduced mutations, * indicates mutations of CCH unique sites. (A-C: Lower) Luciferase activities in cells electroporated with the various chimeric replicons containing introduced mutations, as indicated. The results are expressed as relative luciferase activity. Q, P1059Q; QT, P1059Q plus P1077T; QTV, P1059Q plus P1077T plus D1101V; H, R1276H; HS, R1276H plus D1311S; TV, A2621T plus A2764V; TVN, A2621T plus A2764V plus T2477N.
-
We then proceeded to examine whether the complete CCH NS3 and NS5B regions, incorporating the aa mutations identified could replace their JFH1 counterparts. Five mutations, including P1059Q, P1077T and D1101V identified in pSGR-luc-JFH1/CN3-1, and R1276H and D1311S identified in pSGR-luc-JFH1/CN3-2, were introduced into the CCH NS3 region of the chimeric replicon pSGR-luc-JFH1/CN3. The luciferase assay results indicated that the introduction of these five aa mutations into CCH NS3 was not sufficient to rescue the replication of the chimeric replicon JFH1/CN3 (Figure 5A). Additional candidate aa mutation (T1377I and S1527A) within the CCH unique sites of NS3 were therefore introduced, but these still failed to rescue replication, as indicated by a luciferase expression level that was equal to that of the JFH1/GND replicon (Figure 5A). Instead, therefore, three mutations, A2621T, A2764V and T2477N, were introduced into NS5B and designated as JFH1/CN5 TVN. It was notable that, following the replacement of JFH1 with the CCH NS5A together with the NS5B incorporating these three mutations, the chimeric replicon JFH1/CN5 TVN could then replicate in Huh7.5.1, although at a significantly lower level than JFH1 (Figure 5A). Previously, the isoleucine at position 405 (PMID 22532694) had been identified to be important for the activity of NS5B (Scrima N, et al., 2012). Indeed, when we further introduced this mutation into the JFH1/CN5 TNV constructs, the luciferase activity in cells increased sharply at 72 h post-electroporation, indicating that 405I enhanced replication significantly (Figure 5A).

Figure 5. Luciferase activity and colony formation of NS3-and NS5-substituted replicons. (A: Upper) Schematic structures of the chimeric replicons and the introduced mutations; * indicates mutations of CCH unique sites and # indicates the 405I mutation. (A: Lower) Luciferase activities in cells electroporated with various replicon RNAs, as shown. The results are ex-pressed as relative luciferase activity. (B) Colony formation assay of replicons. Huh7.5.1 cells electroporated with RNAs transcribed from replicons carrying the gene for neomycin phosphotransferase were cultured with DMEM containing G418 for 3 weeks and the cell colonies were stained with crystal violet.
It is possible that the luciferase assay is not sensitive enough to detect low levels of replication. We therefore constructed corresponding replicons carrying the neomycin phosphotransferase gene instead, and colony formation assays were performed in order to determine the replication of the different replicons. Consistent with the results of the luciferase assay, the replicon bearing the complete CCH NS5A and NS5B substitutions but incorporating the "rescue mutations" showed a high rate of colony formation, whereas no colonies were formed in the chimeric JFH1 replicon that incorporated CCH NS3 (Figure 5B). Thus, we demonstrated that the combination of the three mutations A2621T, A2764V and T2477N was essential for CCH NS5B to be able to substitute functionally for the JFH1 NS5B in the JFH1 replicon, but in contrast we failed to find the corresponding amino acids that were responsible for dysfunction in CCH NS3.
-
In order to try to make a replicable CCH replicon with the introduction of the minimum length of sequence from JFH1, we introduced the mutations A2621T, A2764V and T2477N into the pSGR-CCH replicon construct with either a partial or total substitution of the CCH NS3 by the JFH1 NS3 (Figure 6A). To our surprise, the CCH replicon incorporating the entire NS3 from JFH1 could not replicate (Figure 6B), despite the fact that CCH NS4A, NS4B, NS5A and NS5B with mutations of A2621T, A2764V and T2477N individually functioned successfully in the context of the JFH1 backbone. Also, the insertion of the partial JFH1 NS3 into the CCH replicon, together with the mutation of the CCH unique sites in the remainder of NS3 and NS5B, did not result in a replication-competent chimeric CCH replicon (Figure 6B). One possibility for the defective replication of the chimeric HCC might be the low activity of CCH NS5B harboring the mutations (Figure 5A). Indeed, when the 405I mutation was introduced into the CCH/JN3 TNV construct, the construct could replicate at a low level, as shown by the increase of luciferase expression in cells from 72 h post-electroporation (Figure 6B). The final, successful construction of a replication-competent chimeric replicon incorporated the NS3 from JFH1 and four mutations within the NS5B region of the CCH backbone.
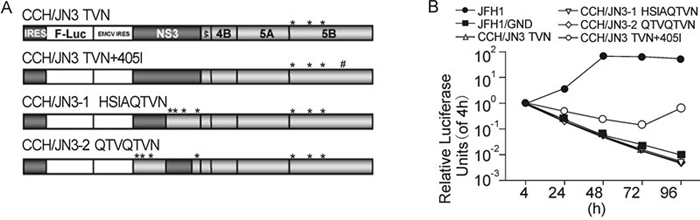
Figure 6. Replication activity of chimeric replicons based on the CCH backbone. (A) Schematic structures of the chimeric replicons and the introduced mutations, * indicates mutations of CCH unique sites and # indicates the 405I mutation. (B) Luciferase activities in cells electroporated with various replicon RNAs, as shown. The results are expressed as relative luciferase activity.
Sequence analysis of the HCV strain isolated from a chronic hepatitis C patient
CCH was replication-defective in the Huh7.5.1 cell line
NS3 and NS5B were responsible for the replication deficiency of CCH
Rescue of the replication of chimeric JFH1 replicons by site-directed mutagenesis
Mutations restore the function of NS5B
Replication-competent chimeric replicons based on the CCH backbone
-
NS3 and NS5B have been reported to be important for the efficient replication of JFH1 (Murayama A, et al., 2007). NS3 is a multifunctional protein that incorporates the proteinase and helicase activity that are involved in both replication and assembly (Ma Y, et al., 2008). The NS3 of CCH shows a total of 43 amino acid differences, relative to JFH1, of which 13 are in the proteinase domain, 29 in the helicase domain, and one lies between them (data not shown). All the key residues that have been identified previously as being required for the proteinase (Yan Y, et al., 1998), NTPase and helicase activities (Frick D N, 2007) are identical between the CCH and JFH1 strains. The NS3 of CCH was functional deficient in the JFH1 backbone; for analysis, it was then divided into three segments with 15, 20 or eight amino acid differences, respectively, relative to JFH1. The replacement of the first two segments abolished replication, but replication could be restored by the separate introduction of either three or two amino acid mutations in the NS3 region. However, the combination of all five amino acid substitutions together in the JFH1/CN3 replicon could not rescue replication. Since most of the reference sequences in the database we used for Blast searching were incapable of replication in Huh7.5.1, other sites besides the CCH unique sites may also be responsible for, or contribute to, the replication deficiency. Another possibility is that the NS3 of CCH, even with the introduction of mutations, may lose the ability to interact with host factors that are important for replication in Huh7.5.1 (Otsuka M, et al., 2005; Ray U, et al., 2011). On the other hand, the CCH unique sites may include residues that contribute to maintaining a high serum copy number in the patient. It was reported that cell-culture-adapted mutations at residues 177 and 254 prevented virus infection in vivo, most likely at the level of interactions with host factors, rather than as a result of defects in the virus itself (Bukh J, et al., 2002). Thus, it is not surprising that a strain with such a high serum copy number during persistent infection in vivo cannot replicate in Huh7.5.1.
NS5B has been identified as a major determinant of efficient replication in JFH1, and the structure and function of the NS5B from JFH1 and J6 isolates have been compared extensively (Schmitt M, et al., 2011; Simister P, et al., 2009). At high resolution, NS5B from JFH1 showed a more closed conformation and this was correlated with a high efficiency of the enzyme in de novo RNA synthesis. The presence of an isoleucine at position 405 stimulates replication in cell culture by enhancing de novo RNA synthesis, and mechanistic analysis shows that I405 promotes both dinucleotide formation and the transition to elongation (Scrima N, et al., 2012). Indeed, in our experiment we also found that 405I in NS5B is important for the replication of CCH strains. Another mutation, at residue 35 (2477N) in the thumb domain, also appears important for replication in Huh7.5.1. This residue is reported to be included in a highly conserved portion that has a stretch that could form a short α-helix, which may be involved in the regulation of open and closed RNA-dependent RNA polymerase conformations (Chinnaswamy S, et al., 2010).
The replication of HCV has been proposed to be driven by the replicase complex, which is composed of the nonstructural proteins, together with several host factors. The direct or indirect interactions between the nonstructural proteins of HCV have been reported to be important for replication efficiency (Dimitrova M, et al., 2003). Recently, Jiang et al (Jiang J, et al., 2012) have shown that adaptive mutations can enhance specific protein-protein interactions amongst viral structural and NS proteins and can therefore promote the assembly of infectious HCV particles. It is possible that mutations in NS4A, NS4B and NS5B influence the interactions between these proteins, which might explain why the substitution of the complete NS4A to NS5B region of CCH led to non-replication (Figure 6B), whereas in contrast the individual substitutions NS4A to NS5A and NS5A to NS5B permitted replication.
Most of the strains recovered from patient serum are reported to be incapable of replication in cell lines but to be able to replicate in primary human hepatocytes or chimpanzees (Buck M, 2008; Yanagi M, et al., 1999). In this study, the CCH strain we isolated from a chronic hepatitis C patient was replication-deficient in cell culture, due to dysfunctions in NS3 and NS5B. The introduction of mutations could partially restore the replication of chimeric replicons, and we finally obtained a replication-competent chimeric construct that incorporated NS3 from JFH1 within the backbone of the CCH strain. Recently, Lu et al. reported a new strategy to generate infectious clones directly from clinical samples by functional screening (Lu J, et al., 2013; Lu J, et al., 2014). An infectious genotype 2a clone was generated that showed 38 amino acid differences compared to the consensus sequence. They found that the majority of these variations were already present in the serum, though in minority. This is consistent with our findings that most of the mutations that we introduced could be found in the clones that were used to generate the consensus sequence. Furthermore, although several subgenomic replicons and HCV cell culture (HCVcc) system have been established, further constructs generated using clinical sequences could facilitate the evaluation of anti-viral drugs.
-
This work was supported by grants from the National Basic Research Priorities Program of China (2013CB911101) and the National Nature Science Foundation of China (Grant 31200315).
-
H Cao and WD Zhu performed the experiments. QX Han participated in the sequence alignment. H Cao and RJ Pei drafted the manuscript. XW Chen conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.













 DownLoad:
DownLoad: