HTML
-
Emerging and re-emerging viral infections threaten hundreds of thousands of human lives every year and cause serious global public health problems. Several well-known viral pathogens include Dengue virus (DENV), West Nile virus (WNV), Yellow fever virus (YFV), Japanese encephalitis virus (JEV) and Tick-borne encephalitis virus (TBE). They are classified into the flavivirus genus in the flaviviridae family (Lindenbach B D, et al., 2007). Some of these virus infections have become increasingly severe, frequently causing epidemics across the world. Taking dengue as an example, there are over 100 million cases of dengue fever occurring in the tropical and sub-tropical regions of the world annually. A number of the cases develop life-threatening conditions, such as dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS) (WHO, 2009). Despite global efforts to develop therapeutics, there are no effective antiviral drugs approved against any of these viral infections (Lim S P, et al., 2013). Vaccine development against DENV has also been slow (Sabchareon A, et al., 2012), although there are vaccines available against YFV (Verma R, et al., 2013), JEV (Yun S I, et al., 2013) and TBEV (Rendi-Wagner P, 2008). To facilitate the development of diagnostics, antiviral drugs, and effective treatments, basic research aims to better understand the mechanisms of viral replication, pathogenesis and virulence factors, transmission patterns, and the varying outcomes of host immune response.
Flaviviruses are positive-sense, single-stranded RNA viruses (Lindenbach B D, et al., 2007). The virus particle has an icosahedral shape and is enveloped, with a diameter about 40–60 nm (Lindenbach B D, et al., 2007). The flavivirus genome is about 11 kilobases and has a 5' end cap similar to that of the cellular mRNA but no 3' end polyadenylation tail (Figure 1A) (Lindenbach B D, et al., 2007). There are also long untranslated regions at both ends, which play important roles in virus replication, viral protein translation, and virus particle assembly (Alvarez D E, et al., 2006; Filomatori C V, et al., 2006; Markoff L, 2003; Polacek C, et al., 2009; Villordo S M, et al., 2009; Yu L, et al., 2008). Viral protein translation results in a single polypeptide precursor spanning across the endoplasmic reticulum (ER)-derived membrane, which is processed into three structural proteins, namely the capsid protein (C), envelope protein (E), and membrane protein (M) and seven non-structural proteins, NS1, NS2A, NS2B, NS3, NS4A NS4B, and NS5 (Figure 1A) (Lindenbach B D, et al., 2007; Mackenzie J, 2005). Both host ER-derived proteases and viral NS2BNS3 protease specifically cleave the precursor to release the individual viral proteins (Figure 1A) (Lindenbach B D, et al., 2007). The C-terminal region of NS3 is a NTPase/ RNA helicase (Xu T, et al., 2005). NS5 possesses N-terminal RNA methyl-transferase (Egloff M P, et al., 2002; Ray D, et al., 2006) and C-terminal RNA-dependent RNA polymerase (RdRP) activities (Lescar J, et al., 2008; Lindenbach B D, et al., 2007). Together with the viral RNA, viral cofactors, and host cofactors, NS3 and NS5 form the virus replication complex on the intracellular membrane to amplify the viral genome (Lindenbach B D, et al., 2007; Murray C L, et al., 2008; Paul D, et al., 2013). Therefore, functional inhibition of the viral non-structural proteins and disruption of the viral replication complex form the rationale behind anti-flavivirus drug development (Bollati M, et al., 2010; Lescar J, et al., 2008; Lim S P, et al., 2013; Malet H, et al., 2008; Sampath A, et al., 2009).
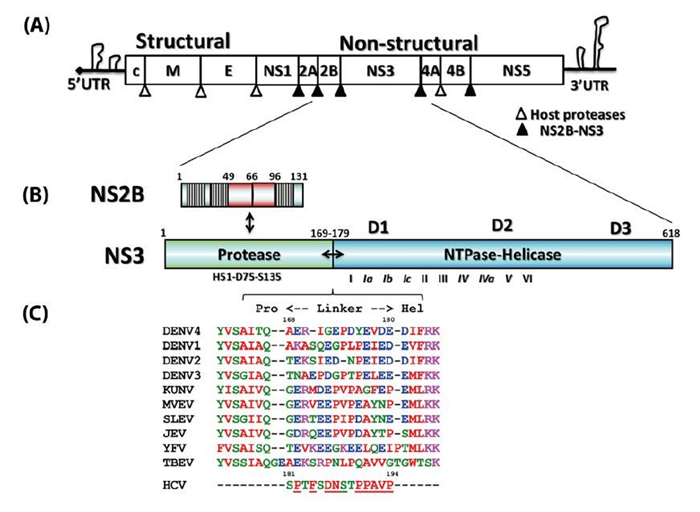
Figure 1. Schematic representation of the flaviviral genome, polyprotein, and functional domain partition along the NS2BNS3 gene. (A) Flavivirus genome and polyprotein. The viral genome contains a 5' cap and untranslated regions at both the 5' and 3' termini. The polyproteins are processed by both NS2BNS3 protease indicated as filled arrows and the host proteases by unfilled arrows. (B) The schematic representation of the NS2B and NS3 genes. The central cofactor region of NS2B is in red, and the putative membrane associate regions of NS2B are displayed as blocks. (C) The sequence alignment of the linker regions between the protease and helicase domains of NS3.
-
Flavivirus NS3 (69 kDa) is the second-largest viral protein (behind NS5) in the flavivirus genome and plays essential roles in the viral life cycle (Figure 1B). NS3 has two functional domains: the N-terminal protease cleaves the viral polyprotein precursor to release individual nonstructural proteins, and the C-terminal's nucleoside-triphosphatase (NTPase)-dependent RNA helicase activity is involved in genome replication and viral RNA synthesis (Lescar J, et al., 2008; Lindenbach B D, et al., 2007; Luo D, et al., 2012). In addition, a recent report described the ATP-independent RNA annealing activity of the DENV NS3 helicase (Gebhard L G, et al., 2012). The two domains are connected by a flexible linker that is about 10 amino acids (aa) in length (Figure 1C) (Luo D, et al., 2010). It is not known why a single gene encodes two distinct domains, but some studies have shown that these two domains are functionally coupled in DENV (Luo D, et al., 2012; Luo D, et al., 2010; Xu T, et al., 2006; Yon C, et al., 2005). Similarly, hepatitis C virus (HCV) NS3 is also a protease-ATPase/helicase fusion protein (Lindenbach B D, et al., 2007) and its activity has also been shown to be regulated by the protease domain (Frick D N, et al., 2004). Recently, several anti-HCV drugs targeting the NS3 protease received U.S. Food & Drug Administration (FDA) approval, and a few promising candidates are undergoing intensive clinical trials (Salam K A, et al., 2013). This will further stimulate the development of antiviral drugs that target the flavivirus NS3 protein. Therefore, we present the recent structural and functional studies of this multifunctional viral enzyme from the flavivirus genus and compare it with the NS3 protein of HCV belonging to hepacivirus genus.
-
The N-terminal domain of NS3 (aa 1–169) is a chymotrypsin-like serine protease and is able to perform both cis-and trans-viral polyprotein cleavage (Chambers T J, et al., 1990; Gorbalenya A E, et al., 1989; Li J, et al., 2005). To function as an active enzyme, NS3 protease requires the cofactor NS2B (Falgout B, et al., 1991; Jan L R, et al., 1995; Yusof R, et al., 2000; Zhang L, et al., 1992). The NS2B protein is a 14-kDa integral membrane protein that contains three domains: two putative hydrophobic transmembrane segments at both N and C termini and a central region (aa 49–96, 47 aa) that is the essential cofactor of the NS3 protease (Figure 1B) (Clum S, et al., 1997). It is known that the flavivirus NS3 protease domain is not very soluble or catalytically active in vitro. Full-length NS3 is not very soluble either, suggesting that the NS3 protease may not fold properly without the NS2B cofactor (Keller T H, et al., 2006; Xu T, et al., 2005). Indeed, Leung et al. engineered a fusion gene of the NS2B cofactor region that was connected to the NS3 protease domain via a flexible nine aa (Gly4-Ser-Gly4) linker; and this fusion protein became soluble when expressed in bacteria and was catalytically active in vitro (Leung D, et al., 2001). This idea of protein engineering greatly facilitated subsequent research on the NS2B-NS3 protease domain and the full-length proteins and promoted advances in NS3 structural biology (Table 1) (Arakaki T L, et al., 2002; Erbel P, et al., 2006; Lescar J, et al., 2008; Luo D, et al., 2008; Nall T A, et al., 2004; Robin G, et al., 2009). Furthermore, the central cofactor region of NS2B was revealed to have dual functions. The presence of the N-terminus (aa 49–67) from the NS2B cofactor region renders NS3 protease soluble but enzymatically inactive, which indicates that NS2B is responsible for the correct folding of the NS3 protease domain (Erbel P, et al., 2006; Luo D, et al., 2008). The C-terminal cofactor region of NS2B (aa 68–96) forms a conserved β-turn hairpin and binds to the hydrophobic S2 and S3 pockets in the substrate binding site of NS3. This peptide of NS2B directly interacts with the bound substrate or substrate-based-inhibitor, supporting the catalytic role of NS2B (Figure 2AB) (Erbel P, et al., 2006; Noble C G, et al., 2012; Robin G, et al., 2009). Thus, the NS2B cofactor is able to regulate NS3pro activity by stabilizing the correct folding of the core structure and by directly participating in the catalysis of substrate cleavage (Chappell K J, et al., 2008; Erbel P, et al., 2006; Luo D, et al., 2012; Robin G, et al., 2009). This is in contrast to HCV NS3pro, which only requires a short sequence from NS4A (~11 aa) to form the active protease. Furthermore, the NS4A cofactor has been shown to play a structural role but does not participate in substrate recognition or catalysis directly (Kim J L, et al., 1996; Tomei L, et al., 1996; Urbani A, et al., 1998).
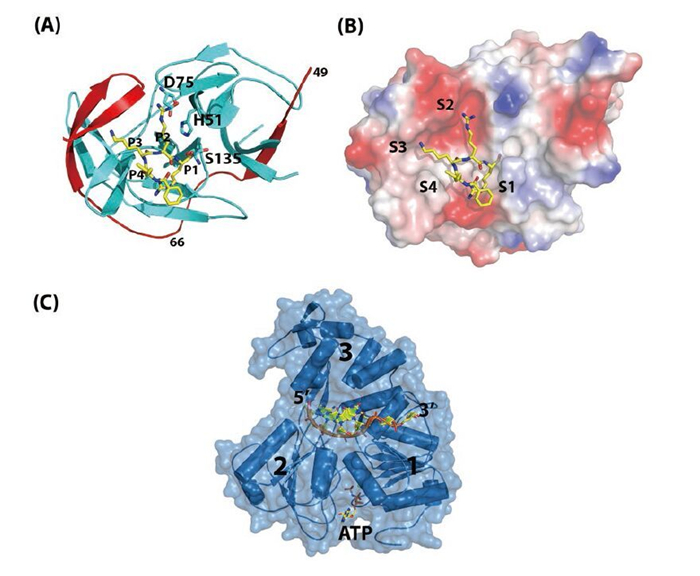
Figure 2. Structures of NS2BNS3pro and NS3hel. (A) Overall fold of NS2B47NS3pro from WNV. NS3Pro is shown in cyan, the NS2B47 region in red, and the peptide inhibitor is in yellow. PDB code: 2FP7. (B) Surface view of the substrate binding site of WNV NS2BNS3pro. (C) Ternary complex of DENV4 NS3hel-ssRNA-AMPPNP. PDB code: 2JLV.
Domains Virus PDB ID Ligands Reference NS2BNS3pro DENV1 3L6P - (Chandramouli S, et al., 2010) 3LKW - DENV2 2FOM - (Erbel P, et al., 2006) DENV3 3U1I Bz-Nle-Lys-Arg-Arg-H (Noble C G, et al., 2012) 3U1J Aprotinin / Bovine Pancreatic Trypsin Inhibitor (BPTI) WNV 2YOL 3, 4-dichlorophenylacetyl-Lys-Lys-GCMA (Hammamy M Z, et al., 2013) 3E90 2-naphthoyl-Lys-Lys-Arg-H (Robin G, et al., 2009) 2IJO Aprotinin (Aleshin A E, et al., 2007) 2FP7 Bz-Nle-Lys-Arg-Arg-H (Erbel P, et al., 2006) 2GGV - (Aleshin A E, et al., 2007) NS3hel DENV2 2BHR SO4 (Xu T, et al., 2005) 2BMF - DENV4 2JLQ - (Luo D, et al., 2008) 2JLR AMPPNP-Mn2+ 2JLU ssRNA12 2JLV ssRNA12, AMPPNP-Mn2+ 2JLW ssRNA13 2JLX ssRNA12, ADP-VO4-Mn2+ 2JLY ssRNA12, ADP-PO4-Mn2+ 2JLZ ssRNA12, ADP-Mn2+ Kokobera Virus 2V6I - (Speroni S, et al., 2008) 2V6J - JEV 2Z83 - (Yamashita T, et al., 2008) MVEV 2V8O - (Mancini E J, et al., 2007) Kunjin Virus 2QEQ - (Mastrangelo E, et al., 2007) YFV 1YKS - (Wu J, et al., 2005) 1YMF ADP NS2B18NS3 DENV4 2WHX ADP-Mn2+ (Luo D, et al., 2010) 2WZQ - 2VBC - (Luo D, et al., 2008) NS2B47NS3 MVEV 2WV9 - (Assenberg R, et al., 2009) Table 1. List of flavivirus NS2BNS3 structures
The protease catalytic triad (His51, Asp75, Ser135) of NS3pro is found in the central cleft (Figure 2AB) (Erbel P, et al., 2006; Noble C G, et al., 2012; Robin G, et al., 2009). In general, the protease recognizes positively charged Arg/Lys at the P1 and P2 positions followed by a small or polar amino acid at P1', although there are exceptions (e.g., Glu is found at the P2 position of the NS2B-NS3 junction) (Gouvea I E, et al., 2007; Li J, et al., 2005; Shiryaev S A, et al., 2007). Fluorogenic peptide substrates are usually used to study protease activities and for screening purposes (Li J, et al., 2005; Niyomrattanakit P, et al., 2006; Yusof R, et al., 2000). Structures of the NS2BNS3pro in complex with substrate-derived peptide inhibitors or aprotinin (bovine pancreatic trypsin inhibitor, BPTI) have revealed the substrate specificity and the catalysis mechanism (Erbel P, et al., 2006; Noble C G, et al., 2012; Robin G, et al., 2009).
NS3 protease inhibitors are currently designed by interfering either with the substrate binding cleft or with the interaction between the NS2B and NS3 protease domain (Lescar J, et al., 2008; Lim S P, et al., 2013). Despite structural and biochemical information available on the NS2BNS3pro substrate binding pocket, no compound has progressed to the preclinical stage to date. Researchers are still facing multiple challenges in identifying effective drugs that target NS2BNS3pro. The substrate binding pocket is shallow and solvent exposed, rendering interactions between mimics and enzyme labile. Moreover, dibasic residues at the P1 and P2 positions hinder peptidomimetic permeability and stability. More recently, researchers began to study the structure and function of the native-state-like NS2BNS3pro in the membrane context, which could lead to better screening platforms for protease inhibitors (Choksupmanee O, et al., 2012; Huang Q, et al., 2013).
-
The C-terminal domain of the multifunctional NS3 protein (aa 180–618) was identified as a member of the superfamily 2 (SF2) helicase (Fairman-Williams M E, et al., 2010; Gorbalenya A E, et al., 1993). The overall structure comprises three subdomains (Figure 2C). Domain 1 and 2 adopt the RecA-like fold (Rao S T, et al., 1973; Story R M, et al., 1992) and contain eight conserved motifs that are essential for RNA binding and ATP hydrolysis activities and their coordination (Fairman-Williams M E, et al., 2010; Lescar J, et al., 2008; Pyle A M, 2008). The third subdomain is unique to help form the single-stranded RNA binding groove. There is also evidence suggesting that domain 3 mediates the interaction with NS5 (Brooks A J, et al., 2002). In addition, NS3 also has RNA 5' triphosphatase activity (RTPase), which shares the same active site for ATP binding and hydrolysis (Figure 2C) (Balistreri G, et al., 2007). RNA 5' triphosphate hydrolysis is believed to be the first step of viral RNA capping (Decroly E, et al., 2012). Viruses carrying a defective or impaired NS3 helicase gene cannot replicate properly, indicating that NS3 is essential for virus replication (Matusan A E, et al., 2001). However, the exact function of the flavivirus NS3hel in the virus replication cycle is unknown. It is speculated that NS3hel could resolve secondary structures of the genomic RNA, displace transacting protein cofactors, and/or separate the dsRNA intermediate that is transiently formed during the polymerization reaction catalyzed by NS5 RdRP into single-strand form (Malet H, et al., 2007; Yu I M, et al., 2008; Yu L, et al., 2008).
Like most eukaryotic DExx helicase proteins, NS3 recognizes RNA largely in a sequence-independent manner (Fairman-Williams M E, et al., 2010; Luo D, et al., 2008; Pyle A M, 2008). During the duplex RNA unwinding process, one strand is inserted into the RNA binding groove of NS3hel, and the sugar-phosphate backbone makes direct or water-mediated contacts to the helicase residues, whereas the other strand is separated by a hydrophobic β-hairpin protruding from domain 2. This β-hairpin acts either actively or passively as a "helix opener" to disrupt base stacking and stabilize the unwound duplex, while the basic concave region between subdomain 2 and 3 works as "the translocator" by binding the 3' overhang (Buttner K, et al., 2007; Luo D, et al., 2008; Myong S, et al., 2007; Pyle A M, 2008). NS3hel binds to the 3' overhang sequence of the duplex RNA and unwinds in the 3' to 5' direction (Figure 2C) (Benarroch D, et al., 2004; Li H, et al., 1999; Xu T, et al., 2005). NTP hydrolysis provides the energy to power the translocation and unwinding process, although the mechanism coupling these two enzymatic activities remains unclear. Single-molecule kinetic studies and structural biology work on HCV NS3hel collectively suggest that the basic step of translocation and unwinding is one base per ATP hydrolysis (Appleby T C, et al., 2010; Dumont S, et al., 2006; Gu M, et al., 2010; Myong S, et al., 2007). Comparative studies on flavivirus NS3 are not yet available and will be of great interest. To date, several flaviviral NS3hel domain structures have been reported (Table 1), providing the structural basis for its enzymatic activities, including basal and RNA-stimulated ATP hydrolysis activity, binding and translocation on the single-stranded RNA, and unwinding duplex RNA.
Helicase inhibitors could serve as common anti-viral therapy against flaviviruses (Lim S P, et al., 2013; Shadrick W R, et al., 2013). HCV NS3hel has been extensively studied, and compound inhibitors have been reported (Li K, et al., 2010; Maga G, et al., 2005; Salam K A, et al., 2013). However, lack of specificity at the RNA binding site and the NTP binding pocket might cause toxicity, as many cellular proteins carry similar helicase/NTPase activities (Linder P, et al., 2011; Shadrick W R, et al., 2013; Steimer L, et al., 2012). The intrinsic flexibility of motor proteins also makes it difficult to identify high-affinity and high potency compounds, although arguably allosteric inhibition remains an attractive concept for inhibitor design (Luo D, et al., 2008; Pyle A M, 2008; Saalau-Bethell S M, et al., 2012). Interesting and potent NS3hel inhibitors have been reported (Byrd C M, et al., 2013; Mastrangelo E, et al., 2012), but no helicase inhibitors have been approved for clinical trials or usage.
-
We do not fully understand why the two independent enzymatic activities hosted by two protein domains–protease and NTPase/helicase–are assembled into the single NS3 protein from flaviviridae. However, there are potential advantages for viruses to encode such a fusion gene. Polyprotein processing occurs co-and post-translationally in ER-derived intracellular membranes. NS3 may help bring NS4A-NS4B-NS5 in proximity to NS2BNS3pro for the efficient cleavage and release of individual viral proteins. In addition, NS2B anchors NS3 in the membrane, which is a prerequisite for viral replication complex maturation. It has been shown that flavivirus RNA replication is performed inside a vesicular-like compartment made of remodeled intracellular membranes (Miorin L, et al., 2013; Paul D, et al., 2013; Welsch S, et al., 2009). Therefore, NS2B together with other small membrane proteins might provide the membrane plat form for the replicative enzymes (NS3 and NS5) and the viral RNA to assemble into the high-order structures of the flaviviral replication complex (Miorin L, et al., 2013; Paul D, et al., 2013; Welsch S, et al., 2009). Interestingly, YFV NS3 has been suggested to be involved in virus assembly independent of its enzymatic functions. A W349A mutation within NS3hel did not result in viral replication defects. However, no infectious viruses but only capsidless subviral particles could be detected from the mutant virus infected cells (Patkar C G, et al., 2008).
Molecular biologists studied the enzymatic activities of the two domains in the context of full-length NS3. Earlier work compared the enzymatic activities of full-length DENV2 NS3 (without NS2B cofactor) with NS3 helicase (aa171–618) and demonstrated that full-length NS3 has much higher unwinding activity but a lower ATP hydrolysis rate (Xu T, et al., 2005; Yon C, et al., 2005). The affinity of dengue virus serotype 4 (DENV4) NS2B18NS3 to ATP analogs is 10-fold higher than that of the truncated helicase (Luo D, et al., 2008). Intriguingly, no influence of the protease on helicase activity has been observed (or vice versa) in an MVEV NS2B47NS3 construct (Assenberg R, et al., 2009). More recently, the Gamarnik group also reported comparable ATPase and helicase activities for his6-tagged DENV2 NS2B47NS3 and NS3hel, respectively (Gebhard L G, et al., 2012). This discrepancy may be due to the variations in protein constructs, enzyme preparation, and assay setup. Further more, the correct folded protease domain might assist NS3 to select the substrate specificity to RNA, as WNV NS3hel, but not NS2B48NS3, unwinds both DNA and RNA (Chernov A V, et al., 2008). Studying the biochemistry of the native-state-like NS2BNS3 in the membrane context could clarify this issue and shed more light on the roles of NS3 in virus replication (Choksupmanee O, et al., 2012; Huang Q, et al., 2013).
Intriguingly, structural studies of full-length NS3 revealed three distinct configurations that differ from each other in the relative positioning of the NS3pro and NS3hel via a flexible inter-domain linker (Figure 3ABC) (Assenberg R, et al., 2009; Luo D, et al., 2012; Luo D, et al., 2010; Luo D, et al., 2008). We reported two different conformations of the DENV4 NS2B18NS3 protein where the protease domain has rotated by approximately 161 degrees with respect to the helicase domain (Luo D, et al., 2012; Luo D, et al., 2010; Luo D, et al., 2008). The structure of the MVEV NS2B47NS3 presented a third and a more radically different conformation of the NS3 protease-helicase (Assenberg R, et al., 2009). Interestingly, small-angle X-ray scattering (SAXS) experiments showed that both DENV4 NS2B18NS3 protein and Kunjin NS3 adopted similar elongated shapes in solution (Luo D, et al., 2008; Mastrangelo E, et al., 2007). Given the close fit of the crystal structures and the hydrated envelope determined ab initio from SAXS data, it is evident that the flaviviral NS3 protease-helicase enzymes maintain an elongated conformation while the two domains are loosely connected through the linker (Figure 3). In contrast, the HCV NS3NS4A protease-helicase has been shown to adopt a globular conformation (Figure 3D) (Yao N, et al., 1999). In the first crystal structure of the similarly engineered NS4A11NS3 fusion protein of HCV genotype 1b, the protease domain binds the C terminus of the NS3hel, mimicking the post-cis-cleavage state at the NS3-NS4A junction (Figure 3D) (Yao N, et al., 1999). NS4A11NS3 fusion proteins from the same or different genotypes have also been crystallized in the similar globular conformation as apo enzyme (Appleby T C, et al., 2010), in complex with ssRNA and ATP analogs (Appleby T C, et al., 2010), in complex with a macrocyclic protease inhibitor (Schiering N, et al., 2011), and in complex with allosteric inhibitors (Saalau-Bethell S M, et al., 2012). Sequence analysis revealed that the linker (aa 169–179) is less conserved compared to the two functional domains of the flavivirus NS3 (Figure 1C). The flavivirus NS3 linker is also rich in acidic residues (Luo D, et al., 2008). Conversely, the linker region of HCV NS3 (aa 181–194) is very conserved and rich in proline residues (Figure 1C). The sequence divergence between flavivirus and hepatitis virus NS3 seems to correlate with the distinct configurations of the protease-helicase: the flexible and less conserved linker is found in the extended conformations of flavivirus NS3 proteins, and the conserved and proline-rich linker is found in the globular conformations of HCV NS3 (Figure 1C and 3). In flaviviruses, the NS3 linker may have an optimum length and flexibility for polyprotein processing and RNA replication (Luo D, et al., 2010). It will be interesting to assess the impact of the HCV NS3 linker on virus replication. These studies on full-length NS3 facilitate a better understanding of the flavivirus life cycle and the exploration of new avenues for the development of antivirals and vaccines (Figure 4) (Luo D, et al., 2010; Saalau-Bethell S M, et al., 2012).
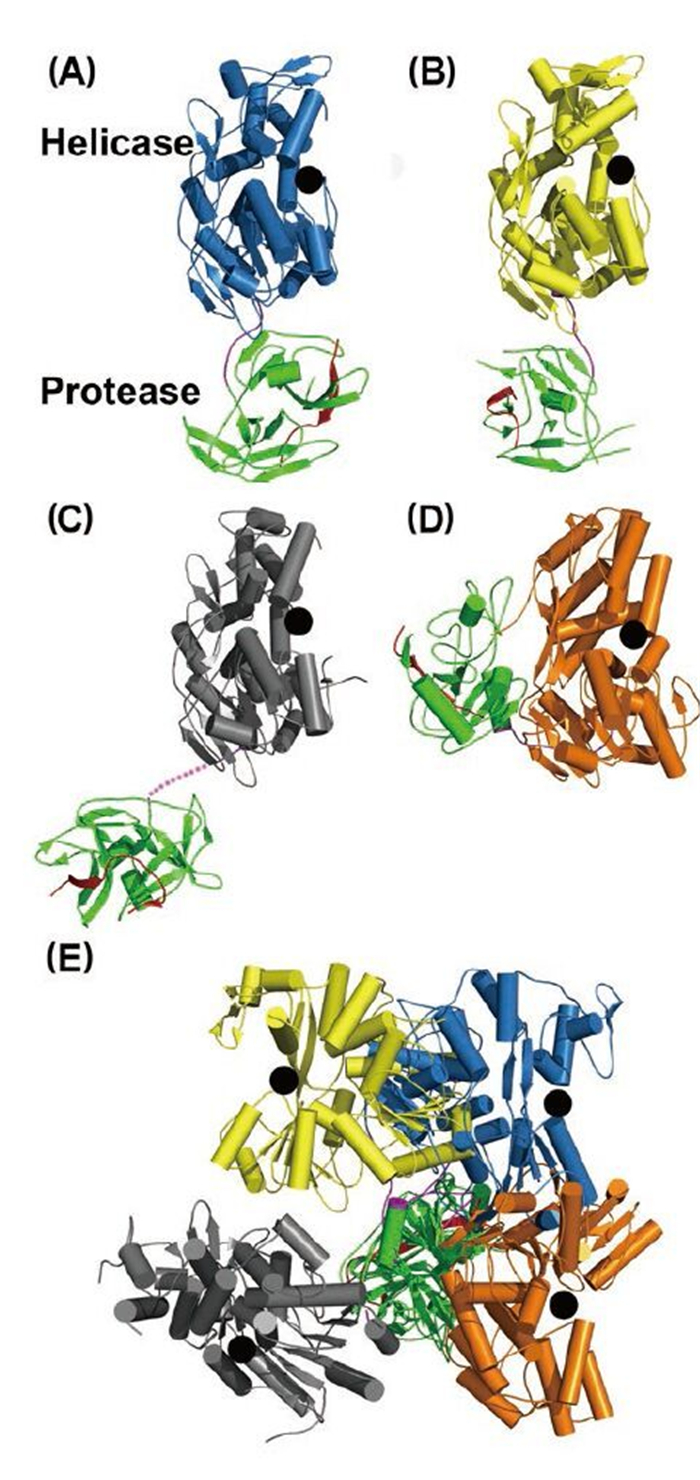
Figure 3. Structural views of the full-length NS3 protein. Structures of (A) and (B) NS2B18NS3 from DENV4, (C) NS2B47NS3 from MVEV, and (D) NS3NS4A11 from HCV. NS2B (or NS4A) cofactor peptide is in red; NS3pro in green; linker in purple; and NS3hel is in blue, yellow, gray, and orange. In (A-D), the helicase domains are aligned, and the black dots represent the ssRNA binding tunnel. (E) When the protease domains are superimposed, the helicase domains display various orientations.
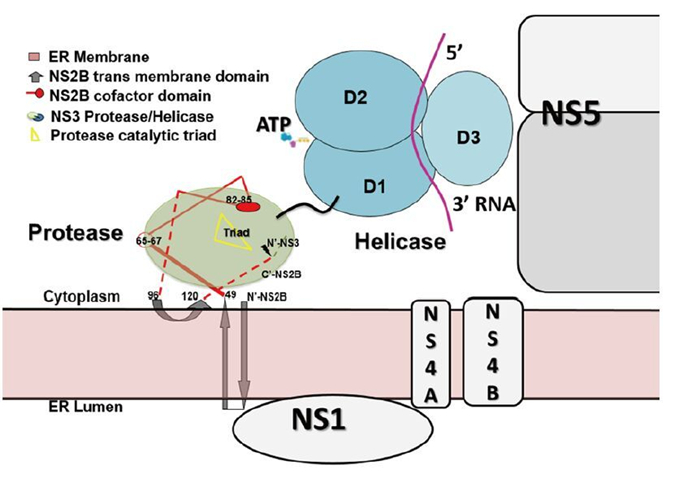
Figure 4. Putative model of the membrane-associated state of the NS2BNS3 complex. The full length of NS2B is modeled based on the predicted topology of NS2B (Luo D, et al., 2010). Numbering of NS2B residues: residues 49–66–96 are the central cofactor region of NS2B colored in red; residues 83–85 directly participate in substrate binding; membrane-associated regions include N-49 and 96-C. NS3pro is colored in green, NS3hel in cyan, and ssRNA in purple. The components of the viral replication complex, including NS5, NS1, NS4A, and NS4B, are drawn across the ER membrane. Lacking information about the precise mapping of the intermolecular interfaces, these individual components are only approximately modeled on the ER membrane.
-
Recent work utilizing yeast two-hybrid systems revealed the flavivirus NS proteins interactome (Khadka S, et al., 2011; Le Breton M, et al., 2011; Zou G, et al., 2011). More than 100 human proteins were identified to interact with NS3, NS5, or both, and many are involved in transcription regulation and host immune response (Krishnan M N, et al., 2014). Type I interferon (IFN) production in human monocyte-derived dendritic cells was inhibited as the cytoplasmic adaptor protein – stimulator of interferon genes (STING) (Ishikawa H, et al., 2008) or mediator of IRF3 activation (MITA) (Zhong B, et al., 2008) – was found cleaved and thus inactivated by DENV NS2BNS3pro (Aguirre S, et al., 2012). WNV NS2BNS3 has been shown to induce apoptosis through the activation of caspases 3 and 8 (Ramanathan M P, et al., 2006). Another study reported that JEV NS2BNS3pro inhibits the signaling pathway of activator protein 1 (AP-1), probably through proteolysis, and the authors suggested that NS2BNS3 protease may contribute to JEV-induced neurotropic pathogenesis (Lin C W, et al., 2006). Better and deeper understanding of how different enzymatic and non-enzymatic activities of NS3 are regulated is needed. The molecular and cellular biology of this protein and its structural and functional roles in viral replication complex formation definitely deserve more attention (Figure 4) as the dynamic nature of the host-pathogen interactions remains unclear (Suthar M S, et al., 2013). Advances in these research areas will inevitably promote the rational design of novel inhibitors and vaccines against diseases caused by flavivirus infections.
-
The authors would like to thank Moon Tay and Julien Lescar for their critical reading of the manuscript.
-
All the authors declare that they have no competing interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.













 DownLoad:
DownLoad: