-
Dear Editor,
Cyanophages are viruses that infect cyanobacteria. They play an important role in shaping the genetic and functional diversity of themselves and their hosts through genetic exchange and shuffling (Lindell et al., 2005; Singh et al., 2012). The psbA gene of cyanophages is a "host-derived" gene, and its encoded D1 protein is capable of maintaining photosynthesis in the infected cells when the host's protein synthesis has been shut down by photoinhibition, thus ensuring a source of energy for phage production (Mann et al., 2003; Lindell et al., 2005). High diversity of viral psbA sequences was observed in both marine waters (Sharon et al., 2007; Chenard and Suttle, 2008; Sandaa et al., 2008) and lake freshwaters (Chenard and Suttle, 2008; Wilhelm and Matteson, 2008; Ge et al., 2013; Zhong and Jacquet, 2013), but the investigations and analysis of cyanophage psbA genes in terrestrial ecosystems have been ignored.
Virus-like particles are more abundant in the paddy water (PW) than those in the ocean (Nakayama et al., 2007). By using culture-independent method, Wang et al. (2009b)firstly obtained 27 different cyanophage psbA sequences from Japanese paddy waters (JPWs), and revealed that the distribution of those sequences was more closely related to that from lake freshwaters than that from marine waters. Given that the biomarker genes g23 of T4-type phages and g20 of cyanomyoviruses in the paddy fields were different between Northeast (NE) China and Japan (Wang et al., 2009a; Jing et al., 2014), we conjectured that novel sequences or clusters of cyanophage psbA genes could be observed in the paddy fields of NE China.
Paddy fields in NE China are irrigated occasionally with river or underground waters, which may impede data interpretation if samplings have been done at inappropriate times. Thus, in this study, an incubation experiment was adopted to survey the psbA genes in paddy waters. Briefly, approximate 10 kg of soil samples (0–10 cm depth) were collected from two paddy fields from DaAn (DA) and MuDanJiang (MDJ) in July 2013 (Table 1). The soils were put into different plastic buckets and irrigated with sterilized water to stimulate the cyanobacterial growth. After thorough homogenization of the soil and water, the buckets were placed outside and supplied with sterilized water daily to maintain approximate 10 cm depth of water layer over soils.
Sample Location Latitude and longtitude Soil type Total C
(g/kg)Total N
(g/kg)Total P
(g/kg)pH Number of psbA clones 5
August20
August4
SeptemberDA DaAn, Jilin 45°36'N,
123°50'ESaline-alka
line soil10.49 0.43 0.37 7.68 70 35 12 MDJ MuDanJiang,
longjiang44°26'N,
129°29'EDarkwn
soil14.99 1.07 0.76 6.27 8 12 23 Table 1. Sampling locations of paddy fields, soil properties and numbers of psbA clones obtained in this study
After 15 days incubation, approximate 400 mL of surface water was collected from each bucket three times in 15-day intervals. The water samples were centrifuged twice at 5, 000 rpm for 30 min at 4 ℃ followed by filtering the supernatant through 0.4-μm and 0.2-μm filters (Whatman Ltd., UK), and viral particles were collected on 0.03-μm polycarbonate membrane filters (Whatman Ltd., UK). The 0.03-μm filters were put into a tube containing 700 µL 10 mmol/L Tris-HCl buffer (pH 7.5), and treated with DNase and RNase (10 µg/mL each) to digest free DNA and RNA. Viral DNA was extracted and PCR amplification was performed with the degenerate primers psbA-F and psbA-R according to the methods reported previously (Zeidner et al., 2003; Wang et al., 2009b). PCR products were cloned and sequenced at the Beijing Genomics Institute (BGI, Shenzhen, China). The sequences were deposited in NCBI database with accession numbers KP729651–KP729662, KP729664–KP729667, KP729669–KP729685, KP729687–KP729723 and KP729725–KP729814.
Totally, we obtained 160 different psbA sequences in this study. Among them 70, 35, 12, 8, 12 and 23 clones were from water samples PW-DA-Ⅰ, PW-DA-Ⅱ, PW-DA-Ⅲ, PW-MDJ-Ⅰ, PW-MDJ-Ⅱ and PW-MDJ-Ⅲ, respectively. The psbA fragments (excluding primers) had lengths of 789–795 bp, encoding 262–264 amino acid residues. BLAST search for the closest relatives at the amino acid level showed that 136 clones were 94%–99% similar to the psbA clones from the JPWs; eleven clones were 97%–98% similar to clones of Synechococcus myoviruses from the East China Sea; four clones were 97%–98% similar to clones from freshwater Lake Kinneret; five clones were 96%–99% similar to isolates of marine Synechococcus phages; four clones (MDJ-Ⅰ-6, MDJ-Ⅰ-7, MDJ-Ⅲ-14, and MDJ-Ⅲ-19) had the highest similarity to uncultured Synechococcus clone and cultured cyanobacteria (Supplementary Table S1).
Phylogenetic relationships among the 160 psbA clones obtained in this study were shown in Supplementary Figure S1. All the clones were roughly grouped into at least seven clusters, of which clusters Ⅰ and Ⅴ were further divided into six and two subclusters, respectively (Supplementary Figure S1). In addition, the GC contents of all clones were in the range of 44.15~ 59.25%, and the triplet peptides of the D1 protein motif R/KETTXXXSQ/H (Sharon et al., 2007) included EEV, EQE, EVE, ENE, ESE, ETE, and EAE (Supplementary Figure S1).
Given that the psbA gene of cyanophage is a "host-derived" gene (Sullivan et al., 2006), problems generally arise on how to distinguish them from their host genes. Phylogenetic tree was a common criterion of distinguishing the environmental psbA sequences coming from phage and host (Sullivan et al., 2006; Chenard and Suttle, 2008). Therefore, all the psbA clones of this study with cyanophage psbA clones from JPWs, marine waters and lake freshwaters, and from isolated cyanophages, as well as psbA sequences from several cyanobacteria such as Synechococcus, Anabeana and Nostoc were subjected to building the phylogenetic tree, and the tree was roughly separated into five major clusters (Figure 1).
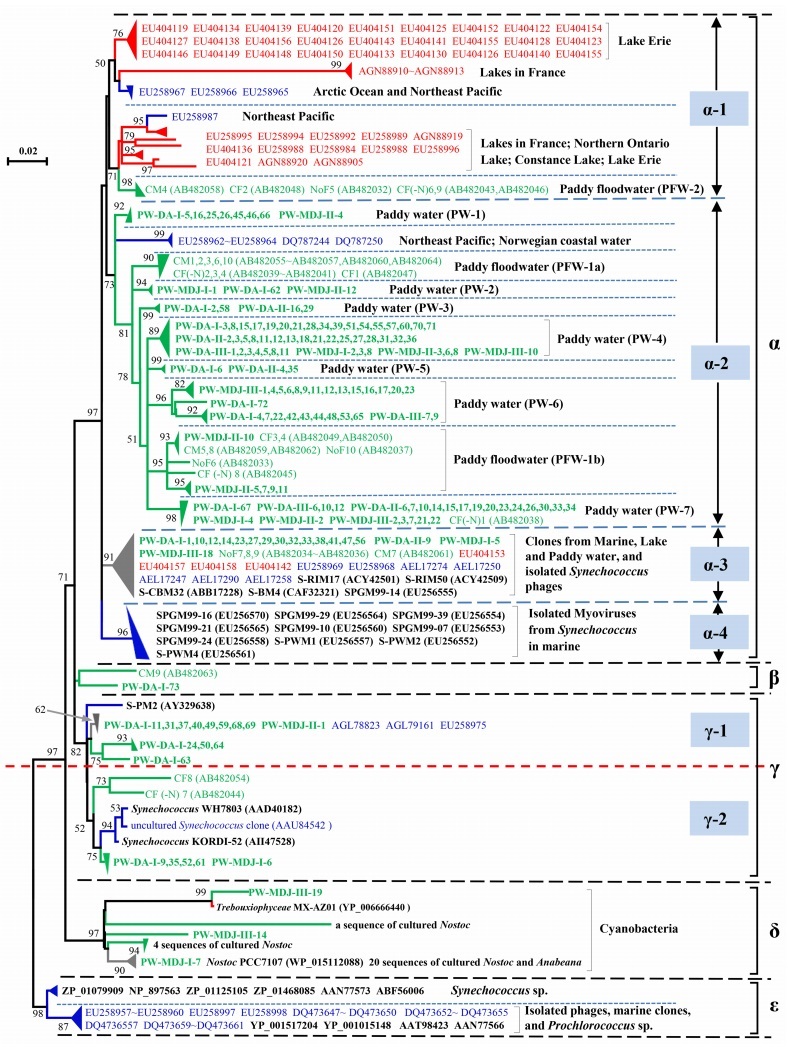
Figure 1. Neighbour-joining phylogenetic tree of 330 psbA sequences, showing the relationships of psbA amino acid sequences from paddy waters in Northeast (NE) China in this study (green words in bold) with those cloned from marine waters (blue words), lake freshwaters (red words), Japanese paddy floodwaters (green words in normal) and obtained from isolated cyanophages and bacteria (black words in bold). Clones in this study were named to reflect sampling sites, sampling data and clone number. In detail, PW stands for paddy water; DA and MDJ respectively stands for DaAn and MuDanJiang; Ⅰ, Ⅱ, Ⅲ stands for sampling date on 5 August, 20 August and 4 September, 2013, respectively. The blue, red and green triangles indicate psbA clusters from marine waters, lake freshwaters and paddy waters, respectively, and the grey triangles represent psbA clusters from more than one environment. The two groups PFW-1 (being further divided into PFW-1a and PFW-1b based on the data of this study) and PFW-2 were previously observed in Japanese paddy floodwaters (Wang et al., 2009b) and seven new paddy water groups (PW-1~PW-7) were designed in this study. Numbers in parentheses are the accession numbers of the psbA sequences from the NCBI website. The clones obtained in this study above the red dotted line were confirmed as phage origins.
Cluster α was the largest cluster, consisting of four subclusters. Approximately 86% (138 clones) of the clones from this study, together with environmental cyanophage clones from marine waters, lake freshwaters and JPWs, as well as isolated phages infecting marine Synechococcus were grouped into this cluster. Cluster β consisted of two paddy water clones from NE China and Japan with 95% similarity between them (Supplementary Table S1). Cluster δ contained three clones obtained in this study and 27 cultured cyanobacteria. We, therefore, concluded these three clones were not from cyanophages. We also found that none of psbA clones from paddy waters fell into cluster ε, which contained isolates of Synechococcus and Prochlorococcus, Synechococcus cyanophage S-SSM1, Prochlorococcus phages, and marine clones. Cluster γ was divided into two subclusters. Subcluster γ-1 contained 13 clones from this study, three marine cyanophage clones and one isolated cyanophage S-PM2. All psbA sequences from paddy waters in this subcluster had viral-like triplet peptides of EQE and ENE, or virus and host like peptide ETE (Sharon et al., 2007). The findings inferred that the psbA sequences in subcluster γ-1 originated from cyanophages. In contrast, subcluster γ-2 contained five clones obtained in this study with three sequences from Synechococcus and two clones (CF8; CF (-N) 7) from JPWs. It should be noted that the origins of clones CF8 and CF (-N) 7 from phages or cyanobacteria were ambiguous (Wang et al., 2009b). Based on the finding of this study, we suggested that the psbA clones in this subcluster might not be from cyanophages (Figure 1). Overall, we isolated 152 different sequences of cyanophage psbA genes from paddy waters of NE China.
The psbA sequences of cyanophages obtained from this study were compared with those from JPWs. In detail, the two groups PFW-1a and PFW-2 consisted of cyanophage psbA sequences only from JPWs, and seven newly designed paddy water groups (PW-1~PW-7) contained clones exclusively from this study, with an exception of a clone CF (-N) 1 from JPWs falling into PW-7. This finding suggested that the psbA gene-containing cyanophage communities in paddy fields in NE China were seemingly different from those in Japan. Notably, since the sequence number was largely unbalanced between the two studies (152 versus 27), we speculated that new insights into phage diversity or distribution would be highlighted as more viral psbA sequences being obtained from paddy waters in Japan or other countries.
Based on the overall structure of the Figure 1, subclusters α-1 and α-2 could be considered as specific clusters for the lake freshwaters and paddy waters, respectively. In addition, both subclusters were split from the same clade, which was far away from the miscellaneous subcluster α-3 containing isolated cyanophages and cyanophage clones from lake freshwaters, marine waters, paddy waters, and from subcluster α-4 containing isolated cyanomyoviruses infecting marine Synechococcus. These findings clearly indicated that the distribution of cyanophage psbA sequences from paddy waters was different from those from lake freshwaters and marine waters.
In conclusion, we successfully isolated 152 cyanophage psbA clones from two paddy waters of NE China. Although the majority of psbA sequences observed in this study had relative high similarity with those from Japanese paddy waters, two and seven specific cyanophage psbA groups were constructed in JPWs and this study, respectively, which suggested that cyanophage psbA assemblages in paddy waters, to some extent were different between two countries. In addition, we also found that the majority of psbA clones from paddy waters of both countries fell into the subcluster α-2, suggesting that the distribution of cyanophage psbA genes in paddy waters is narrow, but far away from those from environmental freshwater and seawater.
HTML
-
This study was financially supported by grants from National Nature Science Foundation of China (41271262), and the Strategic Priority Research Program of Chinese Academy of Sciences (XDB15010103). The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Suppementary figures/tables are available on the website of Virolo-gica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
Clone name
(PW-)Lengtha Alignment Closest relative Source Accession numberb Identity
(%)DA-Ⅰ-1 264 263/264 NoF9 Paddy floodwater in Japan BAH23189 99 DA-Ⅰ-2 264 258/264 CM6 Paddy floodwater in Japan BAH23213 98 DA-Ⅰ-3 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅰ-4 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-5 264 257/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 97 DA-Ⅰ-6 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-7 264 254/264 NoF10 Paddy floodwater in Japan BAH23190 96 DA-Ⅰ-8 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-9 264 256/264 LK20mp-22 Freshwater lake Kinneret (Israel) AAZ93918 97 DA-Ⅰ-10 264 263/264 NoF9 Paddy floodwater in Japan BAH23189 99 DA-Ⅰ-11 264 258/264 PN09-068 Clone in East China Sea AGL79161 98 DA-Ⅰ-12 264 259/264 NoF9 Paddy floodwater in Japan BAH23189 98 DA-Ⅰ-14 264 262/264 NoF9 Paddy floodwater in Japan BAH23189 99 DA-Ⅰ-15 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-16 264 258/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 98 DA-Ⅰ-17 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅰ-19 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅰ-20 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-21 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-22 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-23 264 260/264 NoF9 Paddy floodwater in Japan BAH23189 98 DA-Ⅰ-24 264 248/264 CF (-N) 7 Paddy floodwater in Japan BAH23197 94 DA-Ⅰ-25 264 258/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 98 DA-Ⅰ-26 264 254/264 CM10 Paddy floodwater in Japan BAH23217 96 DA-Ⅰ-27 264 260/264 NoF9 Paddy floodwater in Japan BAH23189 98 DA-Ⅰ-28 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅰ-29 264 262/264 NoF9 Paddy floodwater in Japan BAH23189 99 DA-Ⅰ-30 264 260/264 NoF9 Paddy floodwater in Japan BAH23189 98 DA-Ⅰ-31 264 259/264 PN09-068 Clone in East China Sea AGL79161 98 DA-Ⅰ-32 264 258/264 PN04-091 Clone in East China Sea AGL79481 98 DA-Ⅰ-33 264 260/264 NoF7 Paddy floodwater in Japan BAH23187 98 DA-Ⅰ-34 264 252/264 NoF10 Paddy floodwater in Japan BAH23190 95 DA-Ⅰ-35 264 256/264 LK20mP-22 Freshwater lake Kinneret (Israel) AAZ93918 97 DA-Ⅰ-37 264 258/264 PN09-068 Clone in East China Sea AGL79161 98 DA-Ⅰ-38 264 261/263 Cyanophage
S-RIM17Coastal marine cyanophage ACY42497 99 DA-Ⅰ-39 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-40 264 256/264 DH22-026 Clone in East China Sea AGL78823 97 DA-Ⅰ-41 264 262/264 Cyanophage
S-RIM17Coastal marine cyanophage ACY42497 99 DA-Ⅰ-42 264 254/264 NoF10 Paddy floodwater in Japan BAH23190 96 DA-Ⅰ-43 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-44 264 253/264 NoF10 Paddy floodwater in Japan BAH23190 96 DA-Ⅰ-45 264 252/264 CM10 Paddy floodwater in Japan BAH23217 95 DA-Ⅰ-46 264 257/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 97 DA-Ⅰ-47 264 258/263 Cyanophage
S-RIM17Coastal marine cyanophage ACY42497 98 DA-Ⅰ-48 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-49 264 257/264 PN09-068 Clone in East China Sea AGL79161 97 DA-Ⅰ-50 264 248/264 CF (-N) 7 Paddy floodwater in Japan BAH23197 94 DA-Ⅰ-51 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-52 264 255/264 CF (-N) 7 Paddy floodwater in Japan BAH23197 97 DA-Ⅰ-53 264 254/264 NoF10 Paddy floodwater in Japan BAH23190 96 DA-Ⅰ-54 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-55 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅰ-56 264 259/264 PN04-056 Clone in East China Sea AGL79446 98 DA-Ⅰ-57 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-58 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-59 264 258/264 PN09-068 Clone in East China Sea AGL79161 98 DA-Ⅰ-60 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-61 264 256/264 LK20mp-22 Freshwater lake Kinneret (Israel) AAZ93918 97 DA-Ⅰ-62 264 252/264 CM10 Paddy floodwater in Japan BAH23217 95 DA-Ⅰ-63 264 254/264 CF (-N) 8 Paddy floodwater in Japan BAH23198 96 DA-Ⅰ-64 264 249/264 CF (-N) 7 Paddy floodwater in Japan BAH23197 94 DA-Ⅰ-65 264 254/264 NoF10 Paddy floodwater in Japan BAH23190 96 DA-Ⅰ-66 264 258/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 98 DA-Ⅰ-67 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅰ-68 264 258/264 PN09-068 Clone in East China Sea AGL79161 98 DA-Ⅰ-69 264 257/264 PN09-068 Clone in East China Sea AGL79161 97 DA-Ⅰ-70 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-71 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅰ-72 264 251/264 NoF10 Paddy floodwater in Japan BAH23190 95 DA-Ⅰ-73 264 251/264 CM9 Paddy floodwater in Japan BAH23216 95 DA-Ⅱ-2 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-3 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-4 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-5 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-6 264 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 DA-Ⅱ-7 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-8 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-9 264 261/264 CM7 Paddy floodwater in Japan BAH23214 99 DA-Ⅱ-10 264 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 DA-Ⅱ-11 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅱ-12 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-13 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-14 264 258/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-15 262 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 DA-Ⅱ-16 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅱ-17 264 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 DA-Ⅱ-18 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅱ-19 262 259/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-20 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-21 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-22 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-23 264 259/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-24 264 258/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-25 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅱ-26 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-27 264 256/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅱ-28 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-29 264 257/264 CM10 Paddy floodwater in Japan BAH23217 97 DA-Ⅱ-30 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-31 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-32 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-33 264 259/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-34 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅱ-35 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅱ-36 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-1 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 DA-Ⅲ-2 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-3 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-4 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-5 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-6 264 258/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 DA-Ⅲ-7 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅲ-8 264 259/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-9 264 255/264 NoF10 Paddy floodwater in Japan BAH23190 97 DA-Ⅲ-10 264 254/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 96 DA-Ⅲ-11 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 DA-Ⅲ-12 264 257/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 97 MDJ-Ⅰ-1 264 256/264 Synechococcus
phage S-CBP3Isolation in Chesapeake Bay AFK66481 97 MDJ-Ⅰ-2 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 MDJ-Ⅰ-3 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅰ-4 264 257/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 97 MDJ-Ⅰ-5 264 260/264 LK20mP-12 Freshwater lake Kinneret (Israel) AAZ93917 98 MDJ-Ⅰ-6 264 255/264 uncultured
Synechococcus sp.Transect cruise at eastern AAU84542 97 MDJ-Ⅰ-7 264 262/264 Nostoc sp. PCC7107 Unknown WP_015112088 99 MDJ-Ⅰ-8 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-1 264 258/264 PN09-068 Clone in East China Sea AGL76161 98 MDJ-Ⅱ-2 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 MDJ-Ⅱ-3 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 MDJ-Ⅱ-4 264 259/264 CF (-N) 9 Paddy floodwater in Japan BAH23199 98 MDJ-Ⅱ-5 264 261/264 NoF10 Paddy floodwater in Japan BAH23190 99 MDJ-Ⅱ-6 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-7 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-8 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-9 264 262/264 NoF10 Paddy floodwater in Japan BAH23190 99 MDJ-Ⅱ-10 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-11 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅱ-12 264 254/264 Synechococcus
phage S-CBP3Isolation in Chesapeake Bay AFK66481 96 MDJ-Ⅲ-1 263 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-2 264 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 MDJ-Ⅲ-3 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 MDJ-Ⅲ-4 264 256/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-5 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅲ-6 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-7 264 260/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 MDJ-Ⅲ-8 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-9 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-10 264 260/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅲ-11 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-12 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-13 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-14 264 247/264 Anabaena oryzae
Ind3Unpublished AEZ50165 94 MDJ-Ⅲ-15 264 258/264 NoF10 Paddy floodwater in Japan BAH23190 98 MDJ-Ⅲ-16 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-17 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-18 264 262/264 NoF7 Paddy floodwater in Japan BAH23187 99 MDJ-Ⅲ-19 264 260/264 Trebouxiophyceae
sp.MX-A201Highly acidic Geothermal
LakeYP_006666440 98 MDJ-Ⅲ-20 264 257/264 NoF10 Paddy floodwater in Japan BAH23190 97 MDJ-Ⅲ-21 264 261/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 99 MDJ-Ⅲ-22 264 259/264 CF (-N) 1 Paddy floodwater in Japan BAH23191 98 MDJ-Ⅲ-23 264 254/264 NoF10 Paddy floodwater in Japan BAH23190 96 NOTE: aThe length of amino acid residues; bAccession number of amino acid sequences Table S1. The closest relative of sequenced psbA clones from paddy waters in this study at the amino acid level.
















 DownLoad:
DownLoad: