HTML
-
Highly pathogenic avian influenza (HPAI) H5N1 virus was initially isolated from China in 1996, and termed A/Goose/Guangdong/1/96(Gs/Gd). The descendants of Gs/Gd caused much concern during 1997, when an H5N1 virus outbreak in the Hong Kong Special Admi-nistration Region (HKSAR) of China resulted in 18 human infections and 6 deaths (Webster et al., 2006). Re-emergence of H5N1 was then reported during 2003 and continuing occurrences have been recorded ever since, including outbreaks in migratory birds (Liu et al., 2005; Wang et al., 2008; Hu et al., 2011; Sakoda et al., 2012; Naguib et al., 2015). As of 14 December 2015, transmission of Gs/Gd-lineage H5N1 viruses from avian hosts to humans has been reported in 16 countries worldwide, resulting in at least 844 laboratory-confirmed infections and 449 deaths (http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/). H5N1 viruses are now considered endemic amongst domestic birds in China, Southeast Asia and Africa, and have evolved into numerous Clades and sub-Clades (World Health Organization, 2014). In particular, Clade 2.3.2.1 H5N1 viruses were identified for the first time from a dead Chinese pond heron in HKSAR during 2004, and circulated in wild birds during 2009~2011(Hu et al., 2011; Sakoda et al., 2012). Since 2009, Clade 2.3.2.1 viruses have diversified into sub-Clades 2.3.2.1a, 2.3.2.1b and 2.3.2.1c (WHO, 2014; Bi et al., 2015a).
During 2014-2015, a novel Clade 2.3.2.1c H5N1 vi-rus caused several outbreaks amongst wild or domestic birds in Russia, China (Bi et al., 2015a), The United Arab Emirates (Western Asia) (Naguib et al., 2015), Bulgaria and Romania (Europe), and several African countries (FAO, 2015). In China, there have been at least three outbreaks in Sanmenxia Reservior, Inner Mongolia and Qinghai Lake, resulting in the deaths of thousands of wild birds. Therefore, a systematic analysis of the genetic evolution and relationships between these globally circulating viruses will help efforts to understand, prevent and control the spread of HPAI H5N1 virus.
-
Oropharyngeal and cloacal swabs, tissues and organs from sick or dead birds, and feces from apparently healthy birds were collected for virus isolation. Swabs and tissues were maintained at 4 ℃ in viral-transport medium and sample tubes, respectively. Before inoculation, swabs with viral-transport medium were thoroughly mixed. The tissues and organs were homoge-nized using a Qiagen TissueLyser Ⅱ machine (30 cycles/s; 4 min) in 1-mL cold PBS under sterile conditions. The solid debris in the swab and tissue samples was pelleted by centrifugation at 5, 000 ×g for 10 min. The supernatants were then inoculated in 10-day-old chicken embryos for 72 hours at 37 ℃. Viral RNA from HA titer-positive allantoic fluid was extracted using TRIZOL (Invitrogen) according to manufacturer instructions. All gene segments were amplified using Ex TaqTM DNA polymerase (Takara) with the improved primers (Bi et al., 2015b), and sequenced by the ABI 3730XL automatic DNA analyzer.
-
The gene and amino acid sequences were assembled using DNAMAN (Version 7.0) and BioEdit (Version 7.0.5.2). Representative reference sequences were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov) and GISAID (www.gisaid.org) databases via the online Blast search. Multiple sequence alignment was performed using Muscle (Edgar, 2004). Maximum-likelihood phylogenetic trees were inferred using the RAxML software under the GTRGAMMA model with 1000 bootstrap replicates (Stamatakis, 2014).
-
The GISAID accession numbers for the novel genome sequences of the H5N1 viruses are EPI669753-60, EPI669761-68, EPI669769-76, EPI669777-84, EPI669785-92, EPI669793-800, EPI669801-08, EPI669809-16, EPI669817-24, EPI669825-32, EPI669833-40, EPI669841-48 and EPI669849-56.
Virus isolation and gene sequencing
Genetic and phylogenetic analysis
Gene sequences
-
In May 2015, approximately one hundred black-necked grebes (Podiceps nigricollis) were found dead in Inner Mongolia (Qehan Lake), China. Clinical symptoms, including neurological signs (tremor and opisthotonus) as well as diarrhea were observed in these birds. Various organs, oropharynx and rectal swabs were collected from four diseased or dead birds. Influenza A (H5N1) virus was detected and isolated from these samples, and a total of ten full-length genome sequences of the viruses were obtained.
On 8 July 2015, several great black-headed gulls (Larusi-chthyaetus) were found dead in a village near Qinghai Lake, located approximately 1250 km from Qehan Lake. As of 14 July, a total of more than 2300 birds were found dead in Qinghai Lake, where an HPAI H5N1 virus outbreak had resulted in the deaths of thousands of migra-tory birds in 2005(Liu et al., 2005). The majority of the affected birds in the 2015 outbreak were great black-headed gulls, and these birds were found on a single islet or river estuary in Qinghai Lake. Neurological symptoms were also observed in the infected birds. On 14 July, intestine samples were collected from 11 dead birds, including 8 great black-headed gulls, 2 bar-headed geese (Anser indicus) and 1 great cormorant (Phalacrocorax carbo). Three of the birds were detected to be positive for influenza A (H5N1) virus, and full-length genome sequences were obtained.
-
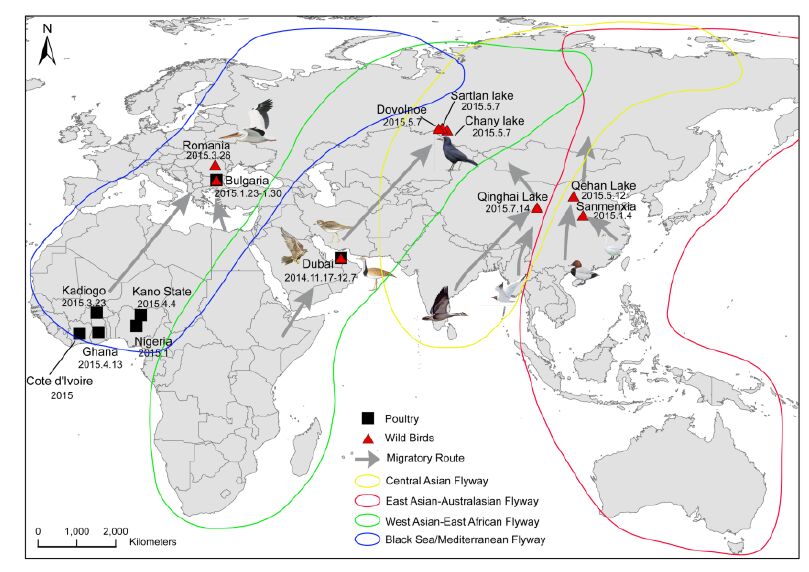
The most closely related sequences for the 13 novel ge-nome sequences were obtained from the GISAID database using BLAST. In addition, the reference sequences for the H5 subtype of HPAI were included for genetic grouping (World Health Organization, 2014). All of the sequences were aligned and phylogenetically analyzed. The HA phylogeny revealed that all of the novel sequences belonged to Clade 2.3.2.1c (Figure 1) and were closely related to sequences isolated from Sanmenxia, China, where HPAI H5N1 viruses caused the deaths of approximately one hundred migratory birds (mostly whooper swans) between late 2014 and early 2015(Bi et al., 2015a). Notably, the Chinese strains also clustered together with those from Russia, West Asia, Europe and Africa that were isolated during May 2015, winter of 2014(Naguib et al., 2015), and spring of 2015(Monne et al., 2015), respectively (Figure 1). For the remaining seven genes, sequences from the strains mentioned above were clustered together and formed an independent sub-Clade (Figure 2 and Supplementary Figures S1-S6), indicating that they belonged to the same genotype. This genotype consisted of a Clade 2.3.2.1c HA (Figure 1) and a PB2 gene derived from H9N2(Figure 2), with the remaining genes originating from other HPAI H5N1 viruses (Bi et al., 2015a; Monne et al., 2015). It should be noted that the prototype virus of this genotype (A/Alberta/01/2014) was first identified from a Canadian patient who traveled to Beijing, China in December 2013(Pabbaraju et al., 2014). However, Clade 2.3.2.1c viruses were subsequently identified from wild birds in the central regions of China, the western regions of Russia, Western Asia and Africa along the Central Asian flyway (Figure 3), suggesting that the novel Clade 2.3.2.1c reassortant viruses are being spread by wild birds along migratory flyways.

Figure 1. Phylogenetic analysis of the novel reassortant Clade 2.3.2.1c HPAI A (H5N1) virus. Maximum likelihood phylogenetic trees of the H5 gene sequences. Sequences isolated from different geographic regions are labeled in different colors.
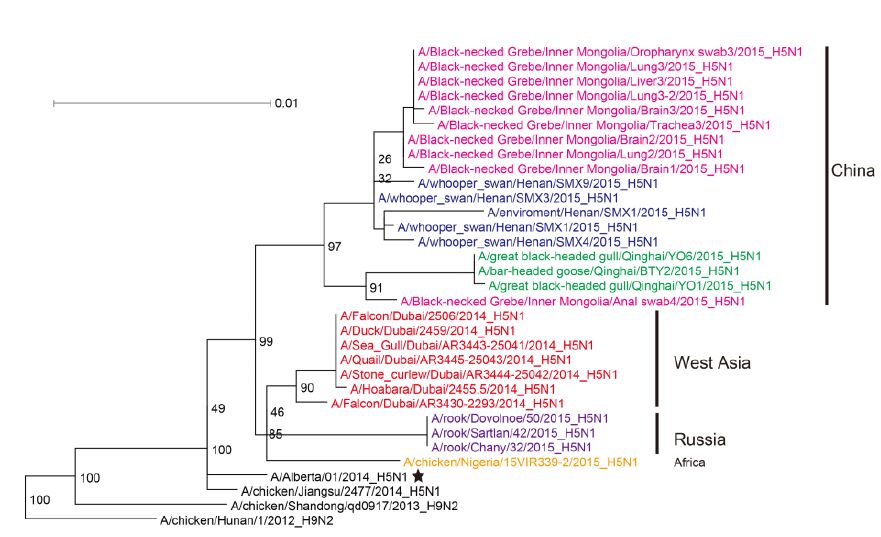
Figure 2. Phylogenetic trees of the PB2 genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
-
The amino acid motif at the HA cleavage site of the H5N1 isolates is RERRRKRG (Supplementary Table S1), indicating that they are highly pathogenic to chickens. In addition, they possess Q226 and G228(H3 numbering) at the receptor binding site, suggesting that they favor avian-like receptors (Stevens et al., 2006). However, they also contain the amino acid substitution T160A, which is thought to enhance the binding capacity to human-like receptors (Zhang et al., 2013). The amino acid deletion at the NA stalk (49-68) is considered a molecular marker for avian influenza virus from aquatic birds for adaptation to land poultry (Matsuoka et al., 2009; Zhou et al., 2009; Cauldwell et al., 2014). Therefore, the Clade 2.3.2.1c H5N1 viruses may have already adapted to poultry before their introduction into migratory birds. Amino acids Q591(Mok et al., 2011), E627(Subbarao et al., 1993; Hatta et al., 2001) and D701(Zhou et al., 2013) in the PB2 protein of the H5N1 viruses suggest that they have not yet adapted to mammalian hosts. No drug-resistance-associated mutations (H274Y in NA; S31N in M2(Hay et al., 1985; Pinto et al., 1992)) were observed among migratory H5N1 viruses, suggesting that these H5N1 isolates are still sensitive to NA and M2 inhibitors.
Disease outbreak, virus isolation and gene sequencing
Phylogenetic analysis
Molecular characteristics
-
Considering that influenza A viruses undergo frequent genetic reassortment, the relatively stable genetic constellation of this genotype during its circulation over long periods of time and distances imply that this novel HPAI H5N1 virus might have gained certain selective advantages that allow for the stable passaging of its viral ge-nome. Clade 2.3.2.1c viruses were identified from wild birds in the central regions of China and the western regions of Russia along the Central Asian flyway (Figure 3). This suggests that the novel Clade 2.3.2.1c reassortant viruses are being spread by wild birds along migra-tory flyways. Therefore, countries located along the East Asian-Australian and Central Asian flyways should be aware of the potential introduction of this reassortant vi-rus, as migratory birds carrying Clade 2.3.2.1 H5N1 vi-ruses have already struck Japan and South Korea in the past (Sakoda et al., 2012). Additionally, other countries should also be on the alert for the potential introduction of this virus into their territories, as exemplified by the introduction of Clade 2.3.4.4 H5N1 viruses into North America and Europe during late 2014 (Jhung and Nelson, 2015; Verhagen et al., 2015). A successful example for outbreak control was the H5N1 outbreak amongst whooper swans in Sanmenxia during January 2015, in which high risk regions for virus transmission along the flyways (such as inner Mongolia) were alerted immediately (Bi et al., 2015a), which helped their preparations and in turn played a considerable role in controlling the outbreak. In addition, human infections with Clade 2.3.2.1c viruses constitute an important reminder of the public health risks from migratory birds carrying H5N1.
-
This study was supported by grants from the National Natural Science Foundation of China (31311120063, 81470096, 31570026, 31471253), the Ministry of Education and Science of the Russian Federation (Chinese-Russian project: RFMEFI61315X0045), the intramural special grant for influenza virus research from the Chinese Academy of Sciences (KJZD-EW-L09), the IDRC-APEIR program (106915-001), and Special Project of Ministry of Science and Technology (2013FY113500). GFG is a leading principal investigator of the NSFC Innovative Research Group (81321063). WFS was supported by the "Taishan Scholar" project of Shandong Province. G.W. is the recipient of a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research (CIHR) and the President's International Fellowship Initiative from the Chinese Academy of Sciences (CAS). We thank the data submitters from the GISAID and GenBank Flu databases for the AIV sequences.
-
All the authors declare that they have no conflict of interests. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
YHB, JJC, WFS and GFG conceived and designed the research. YHB, JJC and WFS performed the experiments. ZJZ, MXL, KS, IS, AS, TLC, GW and YBH performed sample collections. DL, WJL and FML provided helpful suggestions about the study.
Supplementary table/figures are available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
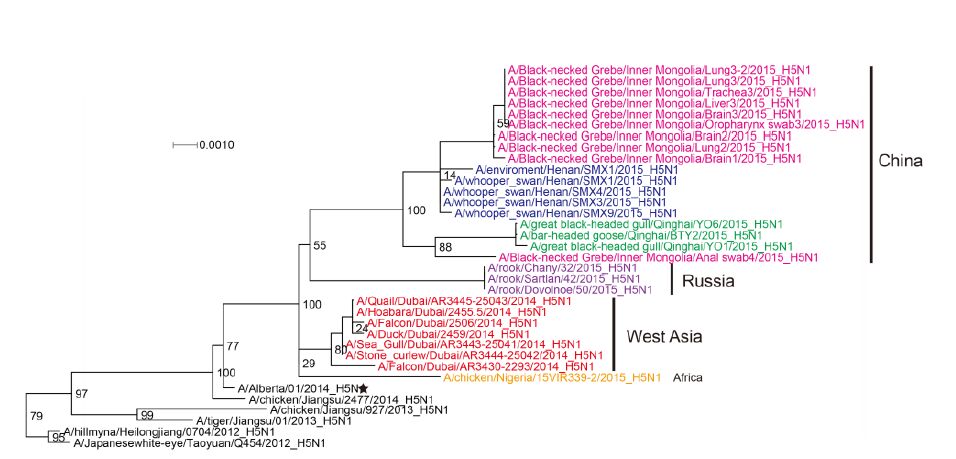
Figure S1. Phylogenetic trees of the PB1 genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1
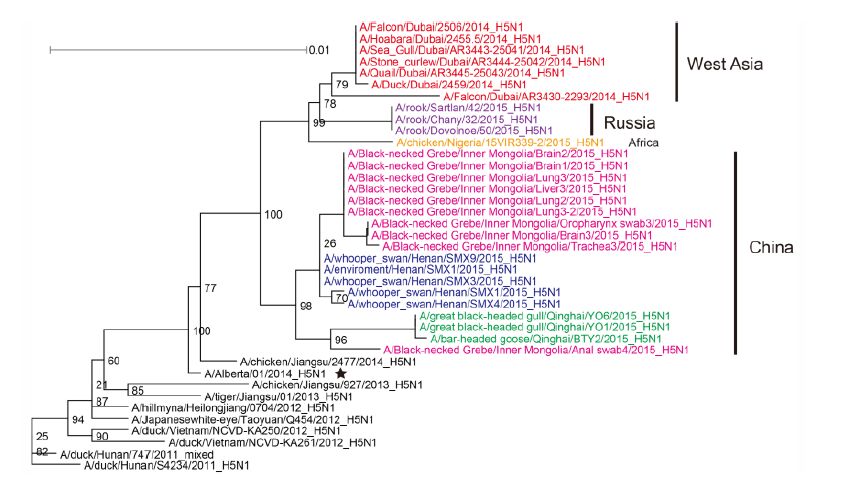
Figure S2. Phylogenetic trees of the PA genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
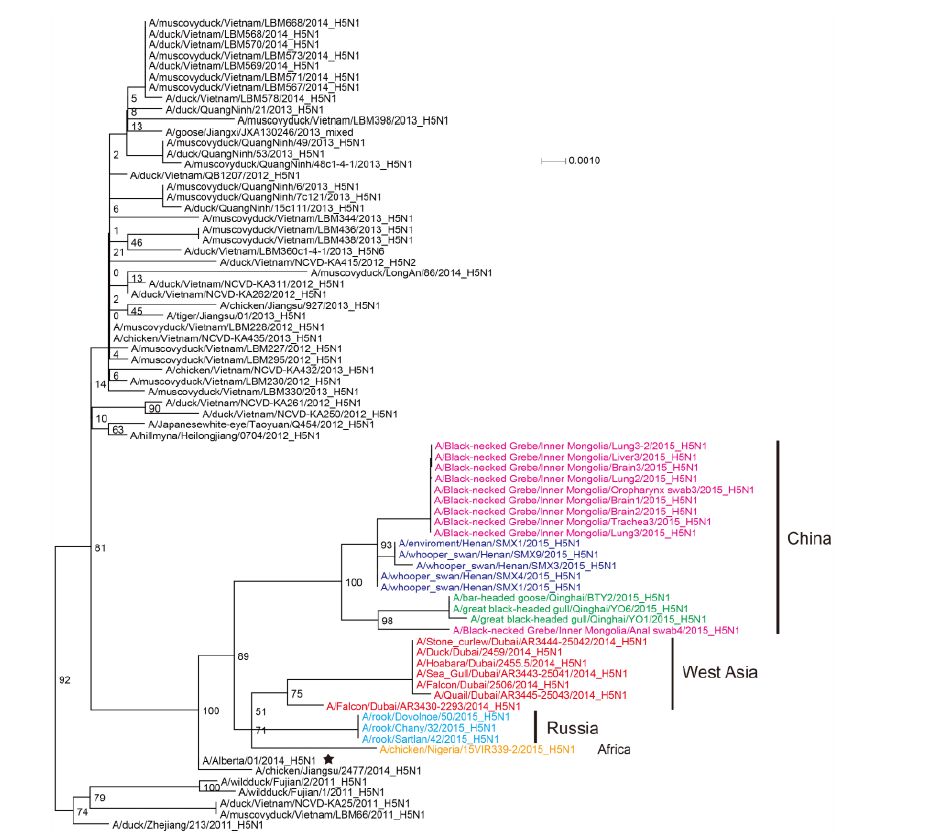
Figure S3. Phylogenetic trees of the NP genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
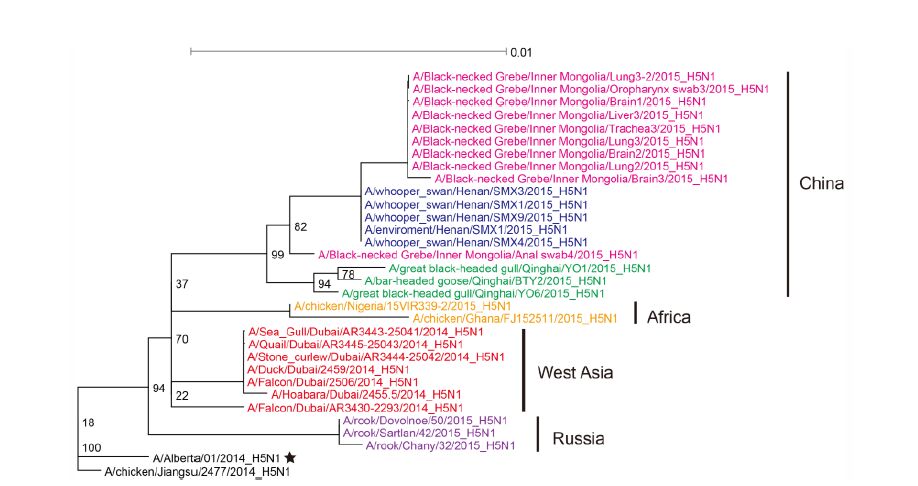
Figure S4. Phylogenetic trees of the NA genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
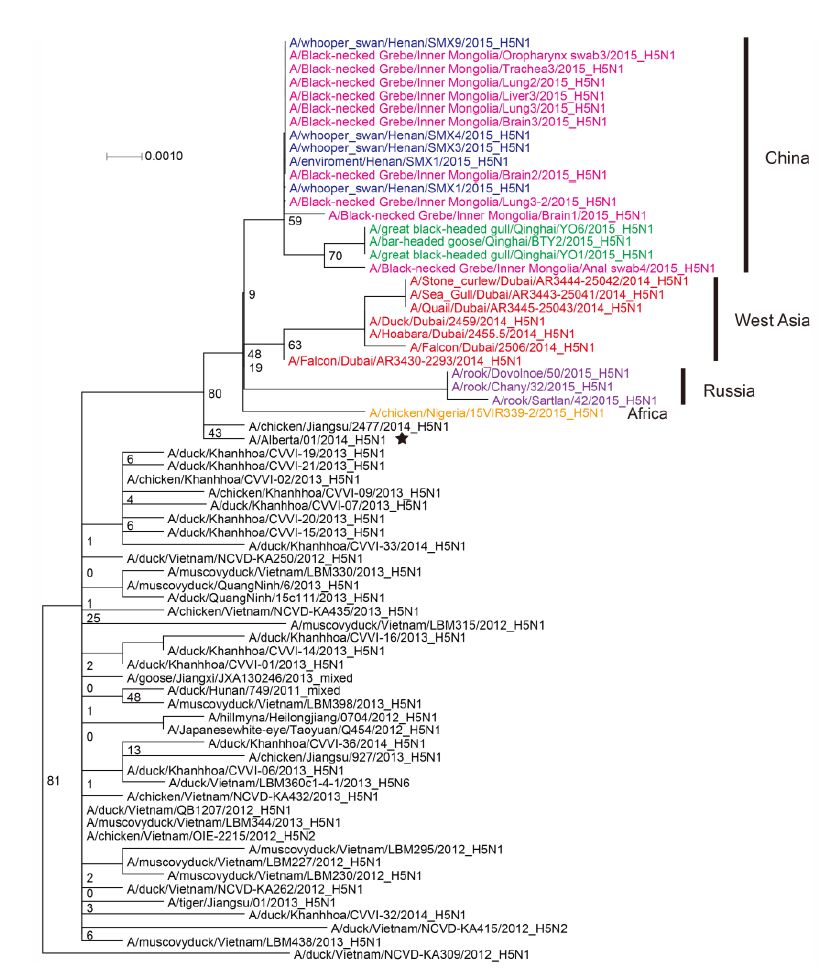
Figure S5. Phylogenetic trees of the M genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
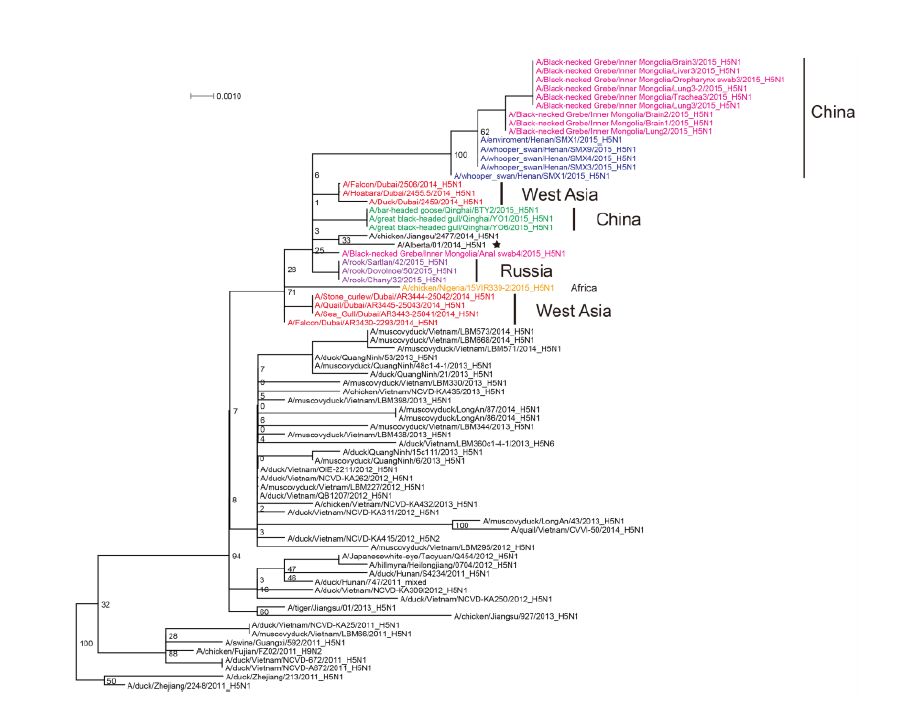
Figure S6. Phylogenetic trees of the NS genes of Clade 2.3.2.1c HPAI A (H5N1) viruses. This figure employs the same coloring strategy as for Figure 1.
Virusesa Collection date HA(H3 numbering) PB2 NA M2 PB1-F2 Connecting peptide 110 158 160 224-228 318 519 627 701 274b Stalk deletion 27 31 58-90 truncated A/tiger/Jiangsu/01/2013 2013.1.20 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Alberta/01/2014 2014.1.3 RERRRKRG H D A NRQSG T Q E D H 49–68 I S Yes A/Stone_curlew/Dubai/AR3444-25042/2014 2014.11.17 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Quail/Dubai/AR3445-25043/2014 2014.11.30 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Sea_Gull/Dubai/AR3443-25041/2014 2014.12.1 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Duck/Dubai/2459/2014 2014.12.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Falcon/Dubai/2506/2014 2014.12.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Falcon/Dubai/AR3430-2293/2014 2014.12.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Hoabara/Dubai/2455.5/2014 2014.12.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/chicken/Ghana/15VIR2588-9/2015 2015(NO DATE) RERRRKRG H D A NGQSG T A/chicken/Ghana/FJ152511/2015 2015(NO DATE) RERRRKRG H G A NGQSG T H 49–68 A/chicken/Nigeria/15VIR339-2/2015 2015.1 RERRRKRG H X A NGQSG T Q E D H 49–68 I S No A/chicken/Niger/15VIR2060-6/2015 2015.4.4 RERRRKRG H D A NGQSG T A/chicken/Burkina_Faso/15VIR1774-35/2015 2015.3.23 RERRRKRG H D A NGQSG T A/duck/Ivory_Coast/15VIR-2742-1/2015 2015.4.13 RERRRKRG H D A NGQSG T A/enviroment/Henan/SMX1/2015 2015.1.5 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/whooper_swan/Henan/SMX1/2015 2015.1.4 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/whooper_swan/Henan/SMX3/2015 2015.1.5 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/whooper swan/Henan/SMX4/2015 2015.1.4 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/whooper_swan/Henan/SMX9/2015 2015.1.5 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/pochard/Henan/SMX1/2015 2015.1.5 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/dalmatian_pelican/Bulgaria/3/2015 2015.1.23 RERRRKRG H N A NGQSG T A/dalmatian_pelican/Bulgaria/4/2015 2015.1.23 RERRRKRG H N A NGQSG T A/chicken/Bulgaria/5407/15 2015.1.30 RERRRKRG H N A NGQSG T A/chicken/Bulgaria/5408/15 2015.1.30 RERRRKRG H N A NGQSG T A/chicken/Bulgaria/5409/15 2015.1.30 RERRRKRG H N A NGQSG T A/pelican/Romania/12449/2015 2015.3.26 RERRRKRG H N A NGQSG T A/rook/Chany/32/2015 2015.5.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/rook/Dovolnoe/50/2015 2015.5.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/rook/Sartlan/42/2015 2015.5.7 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Brain1/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Brain2/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Lung2/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Brain3/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Liver3/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Lung3/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Lung3-2/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Oropharynx swab3/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Trachea3/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Black-necked Grebe/Inner Mongolia/Anal swab4/2015 2015.5.12 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Bar-headed Goose/Qinghai/BTY2/2015 2015.7.14 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Great black-headed Gull/Qinghai/YO1/2015 2015.7.14 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes A/Great black-headed Gull/Qinghai/YO6/2015 2015.7.14 RERRRKRG H D A NGQSG T Q E D H 49–68 I S Yes Table S1. Comparison of key amino acid pattern of Clade 2.3.2.1c HPAI A (H5N1) viruses from different geographic regions
















 DownLoad:
DownLoad: