-
Giant viruses from the Mimiviridae family (Mimivirus, Megavirus, etc.) replicate inside their Acanthamoeba host by the mean of a large intra-cytoplasmic virion factory within which DNA transcription and replica-tion take place using the virus encoded machineries. Members of the Mimiviridae have been isolated in association with much smaller dsDNA viruses called "virophages" that replicate within the virion factory, behaving as parasites of the giant virus. In a recent work Levasseur et al. (2016) used a silencing approach to iden-tify candidate genes possibly involved in the inhibition of the Zami-lon virophage in a specific clade of Mimiviridae. Based on the presence of four copies of a short Zamilon sequence in one of the Mimivirus candidate gene, the authors proposed that resistance to virophages was conferred by a CRISPR-Cas-like system (called MIMIVIRE). Here we dispute this interpretation on the ground that 1) the simultaneous and co-localized replication of the virophage and Mimivirus genomes makes the acquisition of a nucleic acid-based immunity unlikely, 2) the Zamilon-like sequence allegedly acquired by Mimivirus are neither regularly spaced nor flanked by recognizable repeats, 3) the corresponding Zamilon sequence is devoid of distinct flanking sequences that may serve as Protospacer Adjacent Motifs (PAM) discriminating between the virophage and its host. We propose a simpler protein-based interaction model that explains the observed phenomena without having to extend the realm of adaptive immunity to the world of eukaryotic viruses, a revolutionary step that would require stronger experimental evidences.
HTML
-
"Adaptive immunity" qualifies the capacity of an organism to acquire a long-term resistance to a specific infectious agent following a first exposure to it. Whatever its biological implementation, such a system must perform three separate functions: the detection of the foreign status of the "invader", the specific memorization of this encounter, and a mecha-nism inhibiting subsequent re-infections. Adaptive immunity was thou-ght to be the privilege of vertebrates for a long time. Then came the stunn-ing discovery of the CRISPR-Cas system (Goldberg and Marraffini, 2015; Marraffini, 2015) mimicking a similar defense mechanism in prokary-otes. Levasseur et al. (2016) are now proposing that a similar system could operate in certain giant viruses in response to virophage infections. In their recent study, they claimed that 15-nt long fragments of the Zamilon virophage genome once integrated in a specific Mimivirus gene (R349) act as invader-derived "spacers" immunizing the virus against subsequent infections. They tested their hypothesis by showing that silencing the "spacer" -containing R349 gene restored the capacity of the Zamilon virophage to repli-cate in the normally resistant Mimi-virus. Here we review their evidences and conclude that MIMIVIRE is not analogous to the CRISPR-Cas system, does not function via a nucleic-acid recognition system, and is unlikely to possess all the attributes of a bona fide adaptive immune system.
The first and most important difficulty concerns the process of self-nonself discrimination at the root of any adaptive immune system. As far as we know, virophages (La Scola et al, 2008; Gaia et al, 2014) behave as absolute parasites of the virion fac-tory build by Mimivirus within the cytoplasm of the infected Acantha-moeba (Claverie and Abergel, 2009; Mutsafi et al, 2010). This virion factory contains all the necessary enzymatic equipment (most of it encoded by Mimivirus) to transcribe the Mimivirus and the virophage genes (using the same regulatory signals (Claverie and Abergel, 2009; Byrne et al, 2009; Legendre et al, 2010) and replicate their respective genomes at the very same time using a common machinery sequestered in the same compartment. Given the tight co-localization of the two replicating genomes and the absence of significant difference in their G+C contents, the CRISPR-Cas-like mechanism allegedly used by the MIMIVIRE system to discriminate the "invader" from the Mimivirus-own DNA at the initial adaptation stage remains to be conceptualized, if it exists. Leaving the selection of "protospacers" (Goldberg and Marraffini, 2015; Marraffini, 2015) to chance is not an option, as it would lead to the destruction of the progeny of Mimiviruses having selected pieces of their own DNA, regardless of the presence of virophages. To confer any fitness advantage to the Mimivirus host, the putative MIMIVIRE protospacer selection process should be strongly biased toward fragments of the virophage genome. To our knowledge the sole plausible mechanism that has been proposed so far to achieve such feat invokes the preferential selection of protospacer from the most actively replicating DNA molecules, usually corresponding to the small-est "parasitic" genome (e.g. plasmidic vs bacterial) (Wei et al, 2015; Levy et al, 2015). Without the demons-tration that a similar self/non-self discrimination process is at work in Zamilon-infected virion factories, the MIMIVIRE hypothesis and its analogy with an adaptive immune system do not hold.
Even if we admit that MIMIVIRE could preferentially select virophage-derived DNA fragments as CRISPR-Cas-like "spacers", we then run into another difficulty. Once stored within the proposed MIMIVIRE locus (i.e. the Mimivirus R349 gene) as a memory of previous virophage encounters, the spacer itself must escape its targeting at the subsequent interference stage, as such targeting would cause Mimivirus self-destruction. To be effective, the MIMIVIRE defence system must recognize and cut its target when encountered as a unique copy in the Zamilon DNA while sparing it when encountered (in 4 copies) in the Mimivirus DNA molecule. Most of the CRISPR-Cas systems discriminate between the invader-borne protospacers and the cognate host genome-stored "spacer" through the requirement for the so-called Protospacer Adjacent Motif (PAM), a short sequences next to the protospacer recognized by the Cas RNA-guided endonuclease complex (Nishimasu et al, 2015; Yamano et al, 2016). A corollary is then that the PAM short sequence signature must be strictly avoided within the CRISPR locus to prevent self-destruction (Westra et al, 2013; Marraffini and Sontheimer, 2010; Sternberg et al, 2016). This constraint is clearly not obeyed at the MIMIVIRE R349 locus (Figure 1) where 2 copies of the four 15-nt Zamilon-like spacers (highlighted in green) are flanked by adjacent sequences (highlighted in purple) identical or highly similar to those found in the Zamilon genome. This makes a nucleic acid-based discrimination of the Zamilon-borne target from the identical 15-nt spacers integrated at the R349 locus hard to conceive.
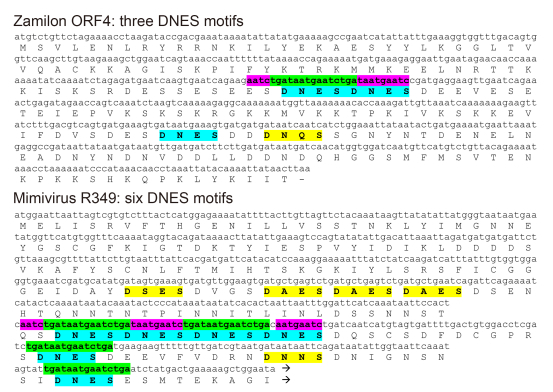
Figure 1. Location of the Zamilon-like subsequence and of the "DNES" amino-acid motifs in Zamilon ORF4 and Mimivirus R349. The 15-nt identical sequences pointed out by Levasseur et al. (2016) are highlighted in green, their identical or quasi-identical flanking sequences in purple. The "DNES" protein motifs are highlighted in blue. R349 exhibits several "DNES" related motifs (e.g. "DAES", "DNNS", "DNQS", "DSES") (highlighted in yellow) in addition to exact copies of the repeats, suggesting that the local similarity with ORF4 might be due to compositional bias and convergent evolution rather than to the transfer of DNA segments. The C-terminal part of the R349 sequence is not shown. In contrast with known CRISPR loci, the "spacer" (highlighted in green) allegedly derived from Zamilon and integrated in the R349 gene are not separated by regularly interspersed palindromic repeats but by "regular" sequences that are not similar to each other and different in length (9 nt, 48 nt, 63 nt).
A final difficulty in comparing the MIMIVIRE process to the CRISPR-Cas system resides in the absence of a well-defined and conserved orga-nization for the storage and presentation of the selected "spacers" Levasseur et al. (2016) assign the role of invader-derived sequence presentation to the R349 gene, within which 15-nt long fragments of the Zamilon genome would have been allegedly integrated, and the silencing of which makes Mimivirus permissive to the virophage replication. However, if the genes orthologous to R349 are virtually identical in all the A-clade Mimiviruses (only 7 of which are publicly available, although 28 were reportedly sequenced) (Levasseur et al, 2016), their homologs in the available B-clade (Megaviruses) and C-clade (Moumouviruses) genomes are highly divergent ( < 35% identi-cal) or severely truncated. In contrast with all previously described prokary-otic CRISPR locus exhibiting a well-conserved organization (the Cas genes, a leader sequence, and a succession of palindromic repeats (Goldberg and Marraffini, 2015; Marraffini, 2015), the R349 locus does not exhibit the hallmarks of a functionally constrained genomic region. Thus, if short Zamilon-like sequences are indeed found in the R349 Mimivirus gene, there is yet no evi-dence that they result from a purpose-ful insertion by a well-controlled adaptive immune response system. Multiple insertions of virophage sequences not targeted to a specific location of host virus genomes have been documented previously (Desnues et al, 2012; Santini et al, 2013). The absence of conserved DNA structures (such as the regularly interspersed palindromic-repeats) presenting the virophage-derived "protospacers" in the Mimivirus ge-nome (Figure 1) and the absence of a conserved region adjacent to the R349 homologs in the other Mimivirus clades argues against MIMIVIRE to be analogous to the CRISPR-Cas system and representative of a bona fide adaptive system capable of conferring immunity to more than a single type of virophage. Additional arguments against MIMIVIRE being analogous to CRISPR-Cas have been presented elsewhere by anonymous authors (URL: https://pubpeer.com/publications/3480B9DE6C9330B0747034C330BA6A).
-
Then if MIMIVIRE is not an evo-lutionary-designed adaptive immune system, how can we reinterpret the results of the silencing experiments described by Levasseur et al. (2016)? First, as noted by the authors, the identical 15-nt DNA sequences shared by the R349 Mimivirus gene and Zamilon's ORF4 are both lo-cated in the respective mRNA sense strands, thus ruling out an inhibitory process mediated by the formation (and degradation) of RNA duplexes. We argued above that a specific defense mechanism triggered by the recognition of the 15-nt repeats at the DNA level appeared unlikely as well in absence of a discriminative signal. We then explored a possible alterna-tive to the nucleic acid-based mecha-nisms proposed by Levasseur et al. (2016). Analyzing the predicted protein sequences encoded by the ORF4 and R349 genes provided several hints that protein-protein interactions could instead be involved in the inhibition of Zamilon replication. We first noticed that the four 15-nt repeats were all inserted in the same translation reading frame. As a consequence, the ORF4 and R349 proteins respectively exhibit 1 and 4 copies of the peptidic motif "Asp-Asn-Glu-Ser" (i.e. "DNES") corresponding to the "tGATAATGAA-TCTga" repeats identified by Levasseur et al. (2016). Interestingly, one additional copy of the "DNES" motif is found elsewhere in the ORF4 protein as well as 2 in the R349 protein (see Figure 1). As it is unlikely that a DNA fragment randomly selected as a putative "protospacer" would coincide with the most frequent 4-amino-acid motif in two unrelated proteins, this raises the possibility that these peptidic motifs, rather than the nucleotide sequences, might be involved in the process of Zamilon inhibition. Accordingly, we find no homolog to the Zamilon "DNES"-containing ORF4 in Sputnik, the virophage able to replicate in Mimivirus (La Scola et al, 2008). Sputnik replication might thus proceed in presence of the "DNES"-containing R349 if this motif does not compete with a protein-protein interaction essential to the virophage infectious cycle. Symmetri-cally, Mimiviridae of the B (e.g. Moumouvirus Monve) and C (e.g. Megavirus courdo7) clades are permissive to the replication of the Zamilon virophage and do not exhibit a "DNES"-containing R349 homolog: the Moumivirus ortholog (mv_R506) to the Mimivirus R349 only shares 29% identical residues and is missing the N-terminal segment containing the "DNES" repeated motifs (Figure 1); the Megavirus ortholog (c7_L652) to the Mimivirus R349 shares 31% identi-cal residues and its N-terminal extremity (1-263) is totally devoid of similarity with R349 and contains no DNES motif. We also did not find any occurrence of the "DNES" motif in the R349 homolog of a newly isolated Megavirus relative produc-tively infected by a close relative of the Zamilon virophage (in preparation).
All of the above results suggest that the coincident presence of the "DNES" motif both in the R349-like protein of a Mimiviridae host and in the ORF4-like protein of a virophage, make the latter unable to replicate. The "DNES" amino-acid sequence is predicted to correspond to a hydrophilic and flexible loop, lo-cated at the surface of the protein. If the successful replication of the Zamilon virophage depends on an interaction of the ORF4 protein with an unknown partner via the "DNES" motif, the presence of the same motif at the surface of the R349 Mimi-virus protein might negatively interfere with this process. Silencing the expression of the R349 protein would then alleviate the inhibition, as observed by Levasseur et al. (2016). To explain the weaker influence of the silencing of R350 and R354 (and the lack of synergy exhibited with the R349 silencing) we propose that these two genes are not specifically involved in the Zamilon inhibition process, but positively influence DNA replication and/or transcription (Figure 2). Their silencing would then allow the replication of the Zamilon virophage by lowering the level of the inhibitory R349, while making the whole replication process less efficient in a pleotropic manner. A testable prediction (although not a definite proof) of our model is that the over-expression of a "DNES"-containing R349 protein in the context of a normally permissive Zamilon-Megavirus system should inhibit the virophage replication. If this is the case, the R349 protein would be a giant-virus-encoded analog to a "restriction factor", i.e. a host protein inhibiting virus (here virophage) infection by interfering with essential virus replication steps such as APOBEC3G with HCV (Zhu et al., 2015), the tetherin paralog oBST2B with various retroviruses (Murphy et al., 2015), or the interferon-inducible transmembrane protein (IFITM) with a variety of pathogenic viruses (Perreira et al., 2013).
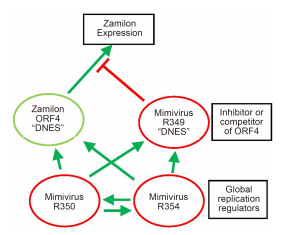
Figure 2. Protein-based interaction model for Zamilon inhibition. The R349, R350, and R354 possess the standard features of bona fide protein coding genes. They are significantly expressed throughout the Mimivirus replication cycle, they exhibit an upstream late-promoter signal and the 3′ downstream hairpin used for mRNA polyadenylation (Byrne et al., 2009; Legendre et al., 2010). We propose that the R349 protein might inhibit the function of the Zamilon ORF4 protein by competing with its normal interactor via their shared "DNES" motifs. Its silencing would then allow ORF4 to perform its essential function, allowing the replication of Zamilon. R350 and R354 might be global replication regulators, the silencing of which might trigger the suboptimal replication of Zamilon by reducing the level of the R349 inhibitory protein. Positive interactions are symbolized by green arrows, inhibitory ones by red "T".
The finding by Levasseur et al. (2016) that Mimivirus could have evolved a specific resistance mechanism against the Zamilon virophage remains fascinating even if it falls short of demonstrating the existence of a CRISPR-Cas-like adap-tive immune system. More studies are needed on different virophage/giant virus systems to assess the existence of a general defense mecha-nism, even before speculating on possible molecular implementations.
-
The IGS laboratory is partially funded by CNRS, Aix-Marseille University, and the French National Research Agency (Grant ANR-14-CE14-0023-01). The authors declare that they have no competing interest. This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: