HTML
-
Mammarenaviruses belong to the genus Mammarenavirus, family Arenaviridae, which consists of 27 unique species currently recognized by the International Committee on Taxonomy of Viruses (ICTV) (Radoshitzky et al., 2015). The original classification of mammarenaviruses, based mainly on virus antigenic properties, serological, genetic and geographical relationships, and the rodent host, identified two groups: New World (NW; also referred to as Tacaribe serocomplex) and Old World (OW; also referred to as Lassa-lymphocytic choriomeningitis serocomplex) mammarenaviruses (Salvato et al., 2005). The NW mammarenavirus are further divided into clades A, B, C and D, in which clade D was once named as a recombinant A/B (recA) clade (Bowen et al., 1997; Clegg, 2002; Emonet et al., 2006). The phylogenetic tree of mammarenavirus recognized by the ICTV, as well as some novel arenaviruses, is shown in Figure 1. The family prototype, lymphocytic choriomeningitis virus (LCMV), was first isolated in 1933 during serial passages in monkeys of human material obtained from a fatal infection in the first documented epidemic of St. Louis enceph alitis (Armstrong and Lillie, 1934; Oldstone, 2007). LCMV is the only mammarenavirus to exhibit a worldwide distribution due to its association with the common house mouse, Mus musculus. Other mammarenaviruses are distributed in North America, South America or Africa. The OW and clade B groups contain the major viruses causing hemorrhagic fevers (HF) on the African continent and in South America, respectively, as well as non-pathogenic viruses. Lassa virus (LASV), Junín virus (JUNV), Machupo virus (MACV), Guanarito virus (GTOV) and Sabiá virus (SABV) are known to cause severe hemorrhagic fever in western Africa, Argentina, Bolivia, Venezuela and Brazil, respectively (Buchmeier et al., 2007). They are included in the Category A Pathogen List as defined by the Centers for Disease Control and Prevention, and are listed as Biosafety Level 4 (BSL-4) agents.
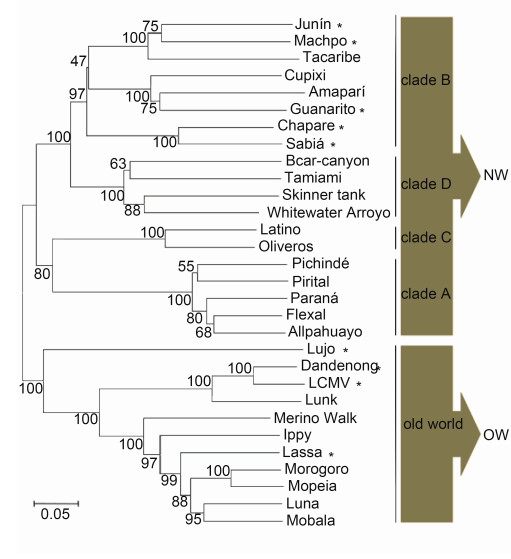
Figure 1. Mammarenavirus phylogenetic tree. Amino acid sequences of mammarenavirus GPCs were aligned using ClustalX. The phylo-genetic tree was computed using the pairwise distance algorithm with neighborjoining ana-lysis in MEGA 4.0 software. Support for nodes was confirmed by bootstrapping using 500 pseudoreplications. The UniProt accession codesincluded are Q90037 (Sabia), Q84168 (Oliveros), Q911P0 (Whitewater Arroyo), Q27YE4 (Ippy), Q9DK03 (Allpahuayo), Q8B122 (Amapari), Q8B121 (Latino), Q8B120 (Parana), Q8B119 (Pirital), Q8B115 (Cupixi), Q8AZ52 (Flexal), Q8AYY5 (Tamiami), Q8AYW1 (Guanarito), Q6IUF7 (Machupo), Q2A069 (Mobala), P26313 (Junm), P19240 (Mopeia), P18141 (Tacaribe), P08669 (Lassa), P07399 (LCMV, lymphocytic choriomeningitis virus), P03540 (Pichinde), G3XEV6 (Luna), D4N7Z6 (Merino Walk), C6ZK00 (Morogoro), C5ILC1 (Lujo), B2KSC1 (Skinner tank), B2C4J0 (Chapare), B1NX58 (Dandenong), A0PJ25 (Bearcanyon) and K0IT71 (Lunk). Skinner tank virus Morogoro virus and Dandenong virus have not yet been recognized by the International Committee on Taxonomy of Viruses. Pathogenic mammarenavirus are indicated by an asterisk.
Mammarenavirus virions are enveloped viruses containing a bisegmented, ambisense RNA genome with four open reading frames. The long (L) RNA segment (≈7, 200 nt) encodes the viral RNA-dependent RNA polymerase (L protein, 200 kDa) and a small RING-finger matrix protein (Z protein, 11 kDa). the short (S) segment (≈3, 500 nt) encodes the nucleoprotein (NP, 63 kDa) and the viral glycoprotein complex (GPC) precursor (75 kDa). The L protein is responsible for transcribing viral mRNAs and replicating the genome (Leung et al., 1979; Fuller-Pace and Southern, 1989; Garcin and Kolakofsky, 1990; Kranzusch and Whelan, 2012). The matrix protein Z plays key roles in budding (Salvato and Shimomaye, 1989; Salvato et al., 1992; Perez et al., 2003; Strecker et al., 2003; Eichler et al., 2004b). The structural NP encapsidates the genome segments and is essential for transcription and replication (Pinschewer et al., 2003).
The GPC precursor is expressed as a polyprotein and matures by undergoing two cleavage events. First, the GPC precursor is cleaved by cellular signal peptidases (SPases) in the endoplasmic reticulum (ER) (Eichler et al., 2003a, b), where the glycoprotein (GP) is extensively Nglycosylated. Unlike most other viruses, the cleaved 58 aa stable signal peptide (SSP) is not degraded but is retained with the immature GP precursor (York and Nunberg, 2007a). The second cleavage event takes place later in the Golgi/trans-Golgi network; the exception is LASV, whose GP is cleaved early in the ER/cis-Golgi compartment (Lenz et al., 2001; Burri et al., 2012; da palma et al., 2014). The cellular proprotein convertase subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P) enzyme recognizes the motif RX(hydrophobic)X ↓ and cleaves the immature GP1/2 into the GP1 and GP2 subunits (Lenz et al., 2001; Beyer et al., 2003; Kunz et al., 2003; Rojek et al., 2008a). In this motif, X is any amino acid except cysteine and the hydrophobic amino acid is preferentially leucine. In OW viruses, the cleavage motif is "RRLX ↓" except for Lujo virus (Pasquato et al., 2011; Burri et al., 2013). The motif present in the LASV GPC is RRLL, which has been proved to be crucial for the cleavage in the ER (Burri et al., 2012). The three non-covalently bound subunits-SSP, GP1 and GP2-form a (SSP/GP1/GP2)3 trimeric complex and traffic to the cell surface for virion budding and release (Young and howard, 1983; Burns and buchmeier, 1991; Eschli et al., 2006) (Figure 2).
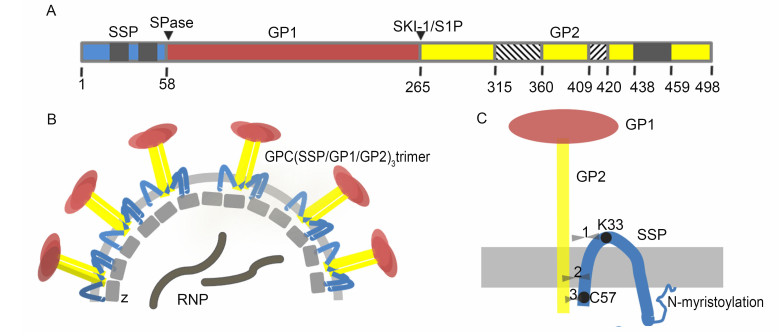
Figure 2. Schematic diagrams of the mammarenavirus envelope GPC. (A) Schematic diagram of the LCMV GPC open reading frame, with the cleavage sites of SPase and SKI-1/S1P indicated. The transmembrane domains in SSP and GP2 are shown in gray. The N-and C-terminal helices in GP2 are shown as lower diagonal and upper diagonal hatching, respectively. (B) Virion structure showing the tripartite GPC, Z protein and ribonucleoprotein (RNP) complex containing NP, L protein and the viral genome. (C) Detailed schematic diagram of the SSP/GP1/GP2 heterotrimer. The three opposing arrowheads represent the potential interaction sites between SSP and GP2, which are discussed in detail in the text. The drawing is not to scale.
The unique SSP/GP1/GP2 heterotrimer of mammarenavirus GPCs is very different from other viruses. The retained SSP plays essential roles in GPC biosynthesis, cleavage and maturation, as well as fusion activity (Eichler et al., 2003a; York and Nunberg, 2006; Agnihothram et al., 2007; Saunders et al., 2007; York and Nunberg, 2007a; Messina et al., 2012; Bederka et al., 2014). GP1 is the distal protein on the virion spike and is responsible for receptor recognition and binding (Cao et al., 1998; Abraham et al., 2010). GP2 mediates membrane fusion after the virions are internalized into acidic endosomes (York et al., 2005; Harrison, 2008; Rojek and Kunz, 2008). According to the sequence alignment results, SSP and GP2 are highly conserved, with amino acid identities of 67.3% and 70.51%, respectively (calculated by DNAMAN software). The alignment of GP1 reveals it is less conserved and the amino acid identity is about 33%. The interactions among the three subunits of the GPC are complex. Recently, the crystal structure of the LCMV GP1/GP2 prefusion complex was reported (Hastie et al., 2016). The extending GP1 N terminus interacts with the C helix, two β-strands and fusion loop from GP2, to form the important conformational epitope that is recognized by most neutralizing antibodies (nAbs) (Figure 3) (Robinson et al., 2016). The interaction of GP1 with the receptor initiates endocytosis. In the endocytic pathway, the acidic pH induces the fusogenic conformational change of GP2 and exposure of the fusion region. Then the fusion region inserts into the cell membrane and GP2 undergoes conformational rearrangements, which is the driving force for membrane fusion (Borrow and Oldstone, 1994). Meanwhile, the interaction between SSP and GP2 is important for pH-induced membrane fusion (York and Nunberg, 2009), but the mechanism is not yet fully understood. Elucidating the molecular mechanisms underlining the structure-function relationship of the three subunits is the key for understanding their function and may facilitate novel avenues for combating virus infections.

Figure 3. Crystal structure of the LCMV GP1 and GP2 prefusion complex. The Protein Data Bank (PDB) accession number is 5INE. (A) Cartoon diagram of LCMV GP1–GP2 interactions. The extending GP1 N terminus, N-terminal loop, β1 and η1 (cyan) interact with the C-terminal helix, two β-strands and the fusion loop from GP2 (purple), respectively. α4 in GP1 (cyan) occupies a cleft (purple) in GP2. (B) Putative epitopes are indicated by light red, green, blue and pink highlights. N-Linked glycans visible in the GP1–GP2 crystal structure are illustrated as cyan ball-and-stick models with the linked asparagine.
-
Mammarenavirus infection is initiated by the binding of GP1 to target cell surface receptors, followed by internalization by endocytosis (Borrow and Oldstone, 1994; Martinez et al., 2007; Vela et al., 2007) and pH-dependent fusion with late endosomes, with the subsequent release of the virus ribonucleoprotein complex into the cytoplasm (Meyer et al., 2002). The first identified receptor for arenavirus was α-dystroglycan (α-DG), a highly glycosylated membrane protein acting as a ubiquitous receptor (Cao et al., 1998). α-DG has been identified as the receptor for both OW and clade C mammarenavirus (Rojek et al., 2007a, b), and O-mannosylation modification of DG was reported to be necessary for LASV binding (Kunz et al., 2005; Hara et al., 2011; Jae et al., 2013). It was shown recently that successful infection of LASV requires a receptor switch from α-DG to a lysosomal protein termed LAMP1 in a pH-dependent manner (Jae et al., 2014). Among the NW clade B mammarenavirus, pathogenic viruses use human transferrin receptor 1 (hTfR1) for entry (Radoshitzky et al., 2007; Flanagan et al., 2008). Non-pathogenic clade B viruses use TfR1 orthologs but are unable to gain cellular entry via hTfR1 (Abraham et al., 2009). However, infection by Lujo pseudotype virus uses neither hTfR1 nor α-DG (Tani et al., 2014). As shown in Figure 4E, Lujo virus GP1 has a low amino acid homology with other OW mammarenaviruses, which might make the receptor-binding sites significantly different from others. Recently, some novel receptor candidates for JUNV, LASV and LCMV have been identified, such as C-type lectin family, dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN), liver and lymph node sinusoidal endothelial calcium-dependent lectin (LSECtin), and two members of the TAM family (Axl and Tyro3) (Shimojima and Kawaoka, 2012; Shimojima et al., 2012; Goncalves et al., 2013; Martinez et al., 2013). Although α-DG and TfR1 are the major receptors, mammarenaviruses may enter into cells in several ways using different molecules, especially in those cells showing low expression of α-DG or TfR1 (Bedossa et al., 2002; Yamamoto et al., 2004).
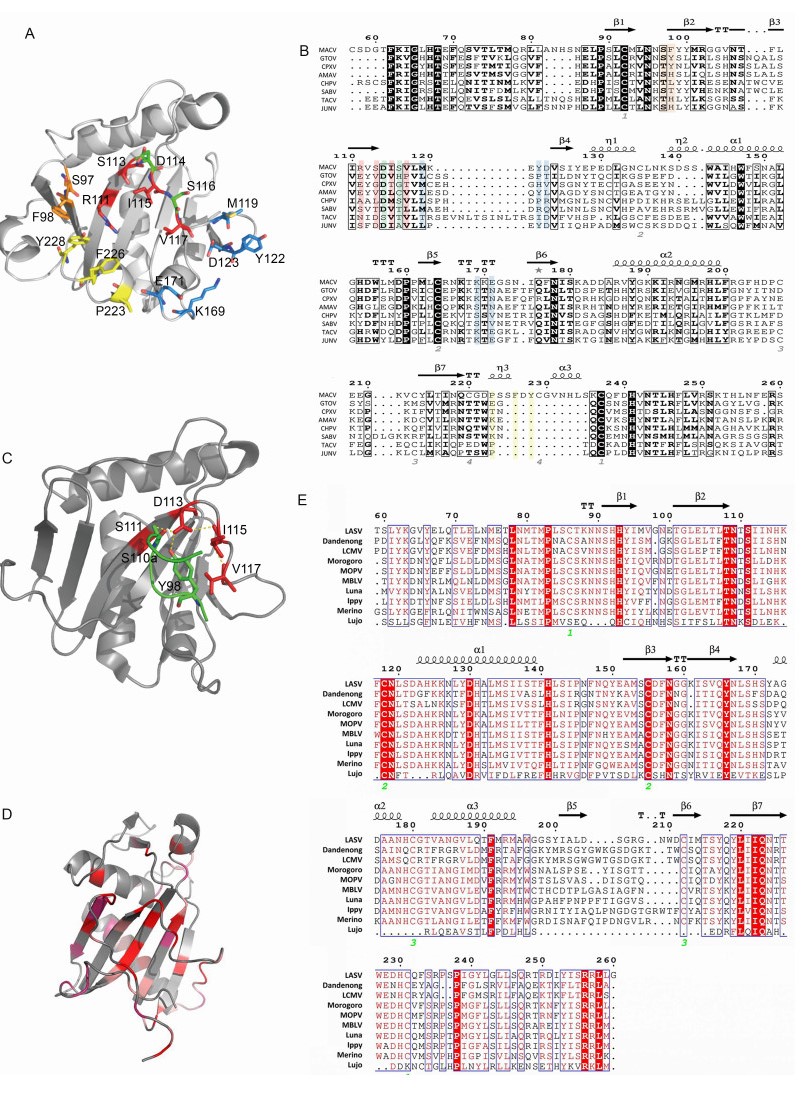
Figure 4. Structure of MACV, JUNV and LASV GP1. (A) The five receptor interaction motifs are shown in red, green, yellow, blue and orange, respectively. (B) Sequence alignment of MACV GP1 aa 77–262 with other NW clade B mammarenaviruses: Chapare virus (CHPV), SABV, Cupixi virus (CPXV), Amaparí virus (AMAV), GTOV, JUNV and Tacaribe virus (TCRV). The observed secondary structure is indicated above the sequence. Disulfide bonds are numbered below the alignment. Residues of the five interaction motifs are colored as in (A). Secondary structure elements are graphically represented by ESPript (Robert and Gouet, 2014). (http://espript.ibcp.fr) (C) Cartoon diagram of JUNV GP1 with the complementarity-determining region of nAb GD01 (residues 97–100 aa) shown as sticks. (PDB code 5EN2). (D) Cartoon diagram of LASV GP1 (PDB code 4ZJF). LASV GP1 is colored by a relative conservation score based on multiple-sequence alignment of OW mammarenavirus GP1 sequences, which was calculated using the ConSurf server (Landau et al., 2005). (E) Sequence alignment of LASV GP1 with other OW mammarenaviruses: LCMV, Dandenong virus, Mopeia virus (MOPV), Morogoro virus, Mobala virus (MBLV), Luna virus, Ippy virus, Merino walk virus and Lujo virus, showing the secondary structure elements as observed in the crystal structure of LASV GP1. Fully conserved residues are highlighted with a red background, and partially conserved residues are shown in red.
The crystallographic structure of MACV GP1 was solved for the unbound protein (Bowden et al., 2009) and for its complex with TfR1 (Abraham et al., 2010). The Protein Data Bank (PDB) code for MACV GP1 is 2WFO (GP1 aa 87–239). MACV GP1 is monomeric when crystallized, with a novel α/β-fold, and is stabilized by four disulfide bonds around the overall folds (Figure 4A, 4B). The relatively conserved disulfide bond presented in MACV, JUNV and Tacaribe virus (TCRV) GP1 reflects the close relationship of these three viruses, as shown in Figure 1 (Bowen et al., 1996; Charrel et al., 2002; Cajimat et al., 2009). The co-crystallization structure of MACV GP1 with hTfR1 has also been depicted (PDB code 3KAS). MACV GP1 interacts with the tip of the apical domain of hTfR1. Five interaction motifs in the interface have been identified, as shown in Figure 4A, 4B (Abraham et al., 2010). Some residues in the motifs, such as R111, Y122 and F226, are essential for GP1-TfR1 interaction, while some residues, such as D114 and S116, are expendable. Besides these five motifs, some residues, such as D155 and P160, have been shown to be important for the GP1-TfR1 interaction in NW clade B mammarenaviruses (Radoshitzky et al., 2011). They may aid in the stabilization of GP1 folding and the GP1-TfR1 interaction. Because clade B contains the major pathogenic mammar enaviruses, these conserved and critical residues might provide a novel perspective for developing broad-spectrum inhibitors against HF.
Identification of the receptor-binding domains (RBDs) is critical for drug development and antibody design (Lalezari et al., 2003; Lazzarin et al., 2003; Dorr et al., 2005; Bossart et al., 2009; Zu et al., 2014). nAb targeting the RBD of filovirus blocks both Marburg virus and Ebola virus entry (Flyak et al., 2015; Hashiguchi et al., 2015), suggesting that RBD is a candidate target for broad-spectrum inhibitor design. The crystal structure of JUNV GP1 bound to the nAb has been reported (PDB code 5EN2; Mahmutovic et al., 2015). The molecular mechanism of antibody-mediated neutralization revealed that the antibody occupied the RBD of JUNV GP1, which was prevented from binding to the cell receptor (Figure 4C) (Mahmutovic et al., 2015).
The crystal structure of LASV GP1 in an acidic environment has been reported (PDB code 4ZJF, GP1 aa 75–237; Cohen-Dvashi et al., 2015). Structure alignment of LASV GP1 and MACV GP1 has revealed that the most structurally conserved region is the central part of the core β-sheet, while highly diversified structures exist at the termini of LASV GP1, which poses a steric barrier that precludes the binding sites contacting TRf1 (Cohen-Dvashi et al., 2015) (Figure 4D). This may reflect the evolutionary distance between OW and NW viruses. However, the reported structure of LASV GP1 obtained at acidic pH could pull down endogenous LAMP1 at pH 5.0 but not α-DG at pH 5.0 or at pH 8.0, indicating that the crystal structure is different from the native structure on the virion spike, which confers a LAMP1-compatible state (Cohen-Dvashi et al., 2015). Although a putative LAMP1-binding motif centered by three histidines (H92, H93 and H230) is conserved in OW viruses (Cohen-Dvashi et al., 2015) (Figure 4E), LCMV has been shown not to require LAMP1 for infection of cells (Jae et al., 2014). The structure of an intact trimeric LASV GPC complex was observed recently by low-resolution electron cryomicroscopy. The tripartite GPC forms the surface spike, which is distributed irregularly on the virion envelope and is connected to the matrix. Following virus internalization, the acidic pH causes GP1 to undergo conformational changes exposing the histidine triad for LAMP1 binding (Li et al., 2016).
The RBD of GP1 bound to α-DG has yet been reported. It has been demonstrated that O-glycan biosynthesis factors, such as the transferase like-acetylglucosaminyltransferase (LARGE), which glycosylates α-DG with O-mannosyl glycans, are indispensable for virus binding (Imperiali et al., 2005; Kunz et al., 2005). Moreover, the interaction of LASV GP1 with LAMP1 is also dependent on the glycans linked to LAMP1 (Jae et al., 2014). Most recently, crystallization of the prefusion LCMV GP1-GP2 complex revealed five critical receptor-binding residues located in or near the basic helix-loop face, which might interact with the acidic sugar on the receptor surface (Hastie et al., 2016). Interesting, in NW viruses such as MACV, GP1 is necessary and sufficient for receptor binding, while in OW viruses such as LASV or LCMV, only the full-length GPC, and not GP1 alone, can achieve α-DG engagement (Jae et al., 2014; Cohen-Dvashi et al., 2015; Hastie et al., 2016). This is similar to the human nAb against LASV. Most human nAbs recognize the conformational epitope consisted of the extended N terminus of GP1 and parts of GP2 that interact with the N terminus of GP1 (Figure 3A) (Robinson et al., 2016).
-
GP2, the membrane fusion subunit of the GPC, belongs to the class Ⅰ viral fusion proteins, presenting a typical α-helix-rich trimer in its postfusion state (Eichler et al., 2006; Klewitz et al., 2007). In the recently reported prefusion structure of LCMV GP2, several striking differences were observed compared with the postfusion structure. In the prefusion state, the long-span N-terminal helix in the postfusion structure was seen as four segments, the T-loop formed antiparallel β-strands instead of the two α-helices in the postfusion structure, and the C-terminal helix formed hydrophobic contacts with the extending N terminus of GP1 (Figure 5A) (Igonet et al., 2011; Hastie et al., 2016). During entry, the gradually acidification triggers a fusogenic conformational change of GP2 (Di simone et al., 1994). The C-terminal helix and N-terminal helix form a postfusion hairpin, which is stabilized by intra-chain salt bridges, and three hairpins gather and form the typical six-helix bundle of a class Ⅰ fusion protein (Figure 5B). The fusion region of LCMV GP2 has a bipartite region that includes an N-terminal fusion peptide and an internal fusion loop, which is thought to be identical across mammarenaviruses.

Figure 5. Structure of mammarenavirus GP2. (A) Cartoon diagram of LCMV GP2 in the prefusion state (PDB code 5INE). (B) Superposition of postfusion GTOV GP2 (gray, PDB code 4C53) with LCMV GP2 (colored as in (A), PDB code 3MKO), generated by the PyMOL Molecular Graphics System, version 1.2r3pre (Schrödinger, LLC). Residues involved in intrachain salt bridges are shown as salmon ball-and-sticks.
In the postfusion state, the inter-chain salt bridge, formed by D353 and R358, and the strictly conserved N325, chelating the central chloride ion, stabilizes the postfusion LCMV GP2 coiled-coil trimer (Igonet et al., 2011). Based on the high amino acid sequence homology, GP2 of OW viruses is suggested to form a conserved structure. Crystallization of the postfusion construct of GTOV GP2 (PDB code 4C53), which belongs to the NW viruses, revealed that it is identical in length to LCMV GP2, with both omitting the fusion region to avoid aggregation, and that they have about 50% amino acid sequence identity and exhibit very similar secondary structures (Parsy et al., 2013) (Figure 5B). Unlike other class Ⅰ viral fusion proteins, GP2 of some pathogenic mammarenaviruses such as LASV and JUNV mediates membrane fusion at a remarkably low pH optimum ( < 4.5 for LASV and < 5.5 for JUNV) (York and nunberg, 2006; Klewitz et al., 2007; Quirin et al., 2008; Rojek et al., 2008b; Cosset et al., 2009; Pasqual et al., 2011). Although the GP2 sequences show high amino acid sequence homology, some minor structural differences might attribute to the difference in sensitivity to acidic pH in late endosomes or lysosomes and the subsequent membrane fusion.
-
Mammarenavirus SSP is essential for virion maturation and subsequent infection (Bederka et al., 2014). As shown in Figure 6, SSP exhibits high amino acid sequence conservation in mammarenaviruses. The 58 aa SSP contains two hydrophobic transmembrane domains (TM1, aa 18–32; and TM2, aa 41–54) linked by a short ectodomain (Figure 2) (Eichler et al., 2003b; Froeschke et al., 2003; Eichler et al., 2004a; Agnihothram et al., 2007; Saunders et al., 2007; Schrempf et al., 2007). SSP plays essential roles in GPC cleavage maturation. SSP association is required in JUNV to allow the exit of the prematuration GPC (pGPC) from the ER and thus for exposure to the SKI-1/S1P protease in the Golgi (Agnihothram et al., 2006). The myristoylated N terminus is assumed to be anchored in the intracellular membrane (York et al., 2004). G2A mutation in the myristoylated motif of LCMV SSP causes reduced virus-like particle (VLP) fusion as well as entry, and blocks GPC-mediated fusion but retains all other SSP functions (Saunders et al., 2007). Similarly, lack of SSP myristoylation in JUNV prevents membrane fusion, with no effect on the formation of the (SSP/GP1/ GP2)3 trimer (York et al., 2008). The penultimate C-terminal C57 is associated with the zinc-finger motif in GP2, which is essential for stabilizing SSP incorporation in GPC (York and Nunberg, 2007b; York and Nunberg, 2007a; Briknarova et al., 2011).
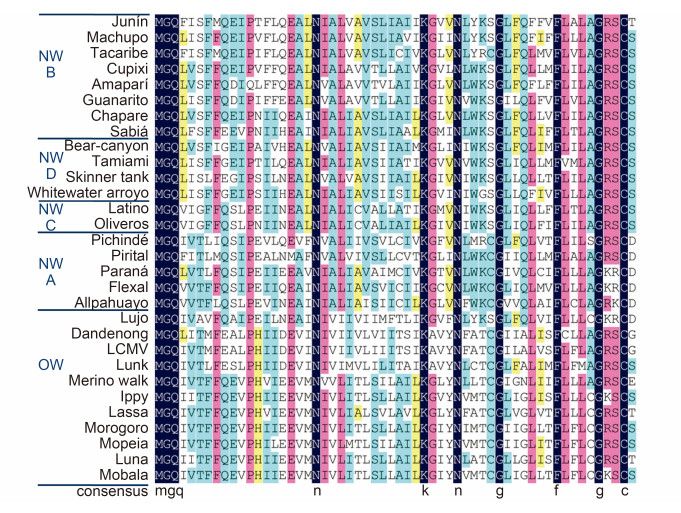
Figure 6. Alignment of mammarenavirus SSP sequences. Sequence alignment was analyzed using DNAMAN, with 100% amino acid identity shown in dark blue, ≥75 in pink, ≥50% in blue and≥33% in yellow. Strictly conserved residues such as the myristoylation motif, K33 and C57 are indicated below the sequences.
Homotypic pairing of the SSP N-terminal cytoplasmic region with the TM1 region (aa 1–32) is paramount for membrane fusion activity. However, homology in the ectodomain, TM2 and the C-terminal cytoplasmic regions appears to be relatively unimportant in fusion activity (Messina et al., 2012). In LASV, pGPC with deletion of either SSP TM1 or TM2 can still traffic to the ER, but deletion of TM2 leads to a reduction of SSP cleavage (Eichler et al., 2004a), which is a prerequisite for GPC maturation (Eichler et al., 2003a). However, somewhat larger TM1 and TM2 deletion mutations (aa 10–32 and aa 34–53, respectively) eliminates virtually all SSP functions.
A conserved hydrophobic motif (FLLL) upstream of the SPase cleavage site is assumed to be the key for GP maturation and trafficking. Mutation of this motif leads to the impairment of GP maturation processing, a reduction of surface expression and the impedance of membrane fusion. Moreover, each of these four residues in this motif has been shown to play an equal role in GPC maturation cleavage and expression. The phenylalanine at position 49 in this motif has also been confirmed to affect both acidic fusion and infectivity (Agnihothram et al., 2007; Saunders et al., 2007).
Some strictly conserved residues across the mammarenaviruses are not only indispensable for SPase cleavage but are critical for SSP function (York and Nunberg, 2007a). A conserved proline at position 12 is essential for infectivity. Both P12A and P12G mutants disrupt VLP fusion and entry. However, a P12G mutant increases membrane fusion activity, while a P12A mutant decreases this, suggesting that SSP plays different roles in GPC-mediated fusion activity and infectivity (Saunders et al., 2007). Moreover, P12 is suggested to be involved in a viral budding motif, ФPXV, which is critical for paramyxovirus budding. In this motif, Ф is any aromatic amino acid and X is any amino acid (Schmitt et al., 2005). The sequence conserved across mammarenaviruses is (I/L/V)PX(hydrophobic residue).
The invariably conserved asparagine residues located at 20 and 37 in SSP have been shown to be essential for GPC function. N20 is critical for VLP fusion and entry, while both N20 and N37 are required for GPC maturation (York and Nunberg, 2006). In LCMV, a K33R mutation retains a wild-type level of pGPC expression and cell surface transport, as well as GPC cleavage and maturation. Positive-charge mutations at position 33 are better than no-charge mutations, and no-charge mutations are still better than the negative-charge mutations for GPC maturation and downstream function. This positive-charge preference at position 33 has also been demonstrated in JUNV SSP (York and Nunberg, 2006). Moreover, K33 in the SSP ectodomain is crucial for GPC sensitivity to acidic pH and subsequent membrane fusion, as discussed above. As indicated above, a K33Q mutant is deficient in membrane fusion. However, mutations in the membrane-proximal external region (MPER) (D400A, E410A, R414A and K417A) in JUNV GP2 were shown to restore this deficiency of pH sensitivity (York and Nunberg, 2009). Hydrophobic residues in the transmembrane domain (F427, W428 and F438) in GP2 also exhibit such complementation, and might interact with residues in the SSP transmembrane domain to stabilize GPC. Interestingly, JUNV K33R, K33H, K33Q and K33E mutations acquire resistance to ST-294, a small-molecule entry inhibitor of NW mammarenaviruses preventing membrane fusion by targeting the SSP–GP2 interface (York et al., 2008).
-
Although the sequence identity of SSP and GP2 in mammarenaviruses is high, hybrid GPC complexes (JUNV SSP with LASV GP1/GP2, or LASV SSP with JUNV GP1/GP2) are unable to induce membrane fusion (Albarino et al., 2011b). However, the hybrid GPC is still correctly assembled and maturation still occurs by cleavage with SPase and SKI-1/S1P (Albarino et al., 2011b; Messina et al., 2012). The mechanism of membrane fusion is not only associated with the conformational changes of GP2 but is also related to the unique SSP subunit, which is much different from other class Ⅰ fusion proteins. The specific SSP and GP2 interaction is essential for stabilizing the prefusion GPC complex. It is reasonable to believe that SSP interacts with GP2 at ectodomain, transmembrane domain and Cterminal cytoplasmic domain (Figure 2C).
Candid #1, a live-attenuated JUNV vaccine, is currently the only vaccine available against an arenavirus and is licensed for human use in Argentina (Maiztegui et al., 1998; Enria and barrera Oro, 2002), Candid #1 has the major attenuating mutation F427I in the transmembrane domain of GP2. This critical residue is suggested to incorporate with SSP to sustain the high-energy GPC trimeric conformation (Albarino et al., 2011a; Droniou-Bonzom et al., 2011; Seregin et al., 2015). Furthermore, York and Nunberg (2007b) identified six conserved histidine and cysteine residues (H447, H449, C455, H459, C467 and C469) in the cytoplasmic domain of JUNV GP2 as essential for the association with SSP. In a later work, the authors demonstrated that H459, C467 and C469 formed a zinc-binding structural domain that incorporated with C57 in SSP to retain SSP in GPC (Briknarova et al., 2011).
The interaction between SSP and GP2 provides a target for drug design against mammarenaviruses. Compound ST-294, showing ability to inhibit NW mammarenavirus, acts through interaction with MPER and transmembrane domain of GP2 (Bolken et al., 2006). Similarly, some mammarenavirus entry inhibitors, such as ST-193, ST-161 and 16G8, demonstrate an antiviral effect by targeting the SSP-G2 interface and perturbing the conformational rearrangement of GPC. The major mechanisms for action of these compounds seem to be stabilization of the prefusion GPC complex (Larson et al., 2008; Lee et al., 2008; York et al., 2008; Cashman et al., 2011). Ngo et al. (2015) reported a compound named F3406 exhibiting strong antiLCMV activity with no cytotoxicity. The target identification revealed that residue M437 within the transmembrane domain of GP2, which has been implicated in SSPGP2 interactions, was critical for virus susceptibility to F3406. Spence et al. (2014) reported a Pichindé virus GP2-derived 19-mer peptide (aa 348–366), AVP-p, with antiviral activity against OW and NW arenaviruses with a 50% inhibitory dose of 7 μM. AVP-p was shown to induce membrane perturbation by changing the lipid organization and affecting the interaction between GPC subunits.
-
Mammarenavirus GPC is a heavily glycosylated protein. It has been estimated that N-linked glycosylation accounts for 40% of the mass of GTOV GP2 (Parsy et al., 2013). N-Linked glycosylation plays an important role in mammarenavirus GPC expression, maturation, receptor binding, membrane fusion and immune evasion (Wright et al., 1989; Braakman and van Anken, 2000; Eschli et al., 2006; Bonhomme et al., 2011; Bonhomme et al., 2013). The GP1 of mammarenaviruses contains 4–11 predicted N-glycosylation sites, in which the positions of potential N-glycosylation sites vary considerably, as shown in Table 1. In contrast to GP1, N-glycosylation in GP2 is well conserved. There are four N-glycosylation sites that are highly conserved in GP2 of mammarenavirus, except for Lunk virus and LCMV, which lack the second N-glycosylation site, and Latino virus, which lacks the third. Interesting, addition of N-glycan in the missing glycosylation site of LCMV GP2 successfully rescued recombinant LCMV, although the virus exhibited mild viral fitness modification toward neurons (Bonhomme et al., 2013). Moreover, some NW mammarenaviruses have a unique N-glycosylation site located in the "top" half of the postfusion crystal structure of the GP2 trimer, while the four conserved sites are located in the "bottom" half (Igonet et al., 2011; Parsy et al., 2013), which might be the result of evolution under selection pressure.
Virus GP1 GP1a GP2 GP2b Length Lujo - 73 - - 93 104 - - 112 - 148 - - - 194 6 - 327 335 352 357 4 454 Dandenong - 80 85 95 - 114 - - 124 - - 171 - - 232 7 - 371 - 396 401 3 498 lcmv - - 85 95 - 114 - - 124 - - 171 - - 232 6 - 371 - 396 401 3 498 Lunk - 77 82 92 - 111 - - 121 - - 168 - - 229 6 - 368 - 393 398 3 495 Merino walk - 73 82 92 - 102 - - 112 122 164 170 - - 230 9 - 371 379 396 401 4 497 OW ippy 66 - 78 88 97 107 - - 117 - 159 165 197 - 228 10 - 369 377 394 399 4 495 LASSA - - 79 89 99 109 - - 119 - - 167 - - 224 7 - 365 373 390 395 4 491 Morogoro - - 78 88 98 108 - - 118 - - 166 - - 222 7 - 363 371 388 393 4 489 Mopeia - - 78 88 98 108 - - 118 - - 166 - - 222 7 - 363 371 388 393 4 489 Luna - - 78 88 98 108 - - 118 - - 166 - - 224 7 - 365 373 390 395 4 491 Mobala - - 78 88 98 108 - - 118 - - 166 - - 224 7 - 365 373 390 395 4 491 NW Oliveros - 75 - 90 101 112 117 - 131 - - 180 - 234 251 9 - 389 397 414 419 4 518 C Latino - 75 - 90 101 112 117 - 131 138 - 180 - 231 248 10 - 386 394 - 416 3 515 Junín - - - 95 105 166 178 - - - 4 - 357 365 382 387 4 485 Machupo - - 83 95 137 166 178 - - - 5 - 368 376 393 398 4 496 Tacaribé - - 83 95 - - - - 124 129 - 176 188 - - 6 - 367 375 392 397 4 495 NW Cupixi - - - - 88 99 - - 125 - - 174 - - 214 5 315 352 360 377 382 5 480 B Amapaí - - - - 88 99 - - 128 - - 174 - - 214 5 315 352 360 377 382 5 481 Guanarito - - - - 88 - - - 125 - - 174 - 202 214 5 314 351 359 376 381 5 479 Chapare - 69 - 88 89 - - - 125 - 171 178 - - 218 7 - 356 364 381 386 4 484 Sabiá - 69 - 88 89 - - - 125 - 171 178 - - 222 7 - 360 368 385 390 4 488 Pichindé 67 74 - 89 100 111 116 121 132 - - 181 217 - 241 11 - 379 387 404 409 4 503 NW
APirital - 75 - 90 101 112 117 122 133 - - 181 218 - 243 10 - 381 389 406 411 4 510 Paraná - 74 - 89 - 111 116 119 130 - - 170 215 219 240 10 - 378 386 403 408 4 507 Flexal - 74 - 89 - 111 116 - 130 - - 179 - 223 240 8 - 378 386 403 408 4 507 Allpahuayo - 74 - 89 - 111 - 119 130 - - 179 - 223 240 8 - 378 386 403 408 4 507 Whitewater arroyo - 73 - 88 - - - 123 126 - 165 176 - - 215 7 315 352 360 377 382 5 480 NW Skinner tank - 73 - 88 - - - - 129 - 168 178 - - 214 6 316 353 361 378 383 5 481 D Tamiami - 73 - 88 - - - 117 128 - - 179 - - 218 6 320 357 365 382 387 5 485 Bear canyon - 73 - 88 - - - - 130 - - 179 - - 216 5 - 355 363 380 385 4 483 Note: a The number of N-Linked glycosylation on GP1. b The number of N-Linked glycosylation on GP2. Table 1. N-Linked glycosylation location in mammarenavirus GPC sequences. Predicted N-linked glycosylation was determined according to NX(S/T) motifs except for NP(S/T) using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/)
Deletion of the first two N-glycosylation sites on GP1 of LASV or LCMV results in a reduction of GPC expression, as well as the inhibition of downstream functions (Schwarz and Aebi, 2011; Bonhomme et al., 2013). The second site is well conserved across NW and OW viruses. According to the MACV GP1 conformation, the pyranose ring of the second N-acetylglucosamine is suggested to form a stacking interaction with F98, which is involved in the interaction with hTfR1 (Bowden et al., 2009; Abraham et al., 2010). In LASV GP1, the second N-glycosylation site (N89) also contributes to building the receptorbinding cluster centered on a unique triad of histidines (Cohen-dvashi et al., 2015). Treatment of MACV GP1 with endoglycosidase F1 makes the deglycosylation GP precipitate, suggesting that N-glycan renders these hydrophobic molecules hydrophilic and thus solubilizes the protein (Parsy et al., 2013). It can be concluded that N-glycan might solubilize the GP and facilitate the receptor recognition and binding.
Moreover, heavy glycan shielding can mask the epitope and promote nAb evasion. Antibody responses against LASV, as well as against other OW mammarenavirus, are not thought to facilitate clearance of viruses, as high viral load and high antibody titers often coexist (Johnson et al., 1987; Gunther et al., 2000; ter Meulen et al., 2000; Fisher-hoch and mcCormick, 2001; Pushko et al., 2001; Meulen et al., 2004). Treatment with human convalescent plasma containing high antibodies titers shows no protective effect against LASV infection (McCormick et al., 1986). Although GP1 shedding and conformational changes were thought to be an evasion strategy (Eschli et al., 2007; Branco et al., 2010; Cohen-Dvashi et al., 2015), a recent study has shown that it was the arenavirus sugar coat that makes the nAb response inefficient (Sommerstein et al., 2015). As shown in Table 1, GP1 of NW clade B viruses have four to seven glycans. It has been reported that there was a negative correlation between nAb response and glycan density in clade B viruses (Sommerstein et al., 2015). JUNV with four glycans in GP1 generates higher nAb titers than viruses with more glycans in their GP1. Second, partially deglycosylated GP1 variants in both NW and OW viruses elicits an accelerated and more potent nAb response to the deglycosylated immunogen (Sommerstein et al., 2015). Meanwhile, antibody induced by wild-type virus neutralizes these specific glycan deletion mutations effectively (Sommerstein et al., 2015). Some key glycosylation sites, such as N99 and N119 in LASV GP1 and N373 in LASV GP2, are suggested to shield the neutralization epitopes, leading to reduction of antibody access to virus (Weber and buchmeier, 1988; Wright et al., 1989; Bonhomme et al., 2013). It has been confirmed recently that most neutralizing human monoclonal antibodies target the linear epitope in GP1 and conformational epitopes in the GP1–GP2 interface, which are located on the glycosylated side that is not fully covered by glycosylation (Figure 3B) (Robinson et al., 2016) Vaccine development for other viruses with such a "glycan shield" provides lessons for mammarenaviruses antibody design. For example, isolation of glycan-binding broadly neutralizing antibodies from human immunodeficiency virus type 1-infected patients strongly highlights the glycan on the envelope GP as a feasible target for vaccine design (Pejchal et al., 2011; Walker et al., 2011; Mouquet et al., 2012; Kong et al., 2013; Garces et al., 2014; Garces et al., 2015).
-
The mammarenaviruses GPC is unique, as SSP retains and forms a heterotrimer with GP1 and GP2. The three subunits then make the (SSP/GP1/GP2)3 homotrimer, which is presented at the surface of the virion and is responsible for cell entry. To date, many questions based on the GPC trimer structure remain unanswered, including dissection of the interaction between SSP and GP2, and its role in virion maturation and membrane fusion, as well as elucidation of the molecular mechanism by which glycans facilitate immune evasion.
The limited treatment strategies against mammarenaviruses make the development of antiviral drugs an urgent need. The high sequence and structural homology of SSP and GP2, as well as use of the same receptor by pathogenic OW or NW viruses, provide novel targets for drug design. Investigation of the structural basis and molecular mechanisms underlying these processes will aid in the development of antiviral therapeutic strategies.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.














 DownLoad:
DownLoad: