HTML
-
HIV-1 is a lentivirus that causes acquired immunodeficiency syndrome (AIDS) (Levy, 1993). Millions of people worldwide are infected with HIV-1 and there is still no cure for AIDS (Quinn, 2008; Archin et al., 2014; Piot et al., 2015). The HIV-1 genome is composed of two copies of positive single-stranded RNA that codes for nine open reading frames (ORFs) that produce 15 viral proteins (Frankel and Young, 1998).
The Gag polyprotein precursor is proteolyzed to four proteins: p17, p24, NCp7, and p6 (Briggs et al., 2004). The Gag-Pol polyprotein precursor is processed into the viral protease (PR), reverse transcriptase (RT, which contains RNA-dependent DNA polymerase, DNA-dependent DNA polymerase, and RNase H domains), and integrase (IN) (Jacks et al., 1988). Env is mainly processed into two proteins, glycoprotein 120 (gp120) and glycoprotein 41 (gp41) (Wyatt and Sodroski, 1998). The other sequences encode auxiliary proteins, including Vpu, Vpr, Vif, Nef, Tat, and Rev (Cullen, 1998).
The HIV-1 genomic RNA, RT, IN, PR, p6, Vpr, Vif, and Nef are enclosed in the core of the virion by a capsid that is composed of p24. The capsid is surrounded by p17. Gp120 and gp41 are associated non-covalently and they are important for viral entry into host cells (Frankel and Young, 1998). The life cycle of HIV-1 can be divided into several distinct steps including viral entry, reverse transcription, pre-integration transcription, integration, transcription, assembly, and budding. Each of the 15 viral proteins plays a role in the life cycle and replication of the virus (Tang et al., 1999; Barre-sinoussi et al., 2013). Protein-protein interactions play a large role in the function of the proteins during the virus life cycle (Swanson and Malim, 2008; Tavassoli, 2011). Thus, research on protein-protein interactions is important in order to understand the life cycle and pathogenesis of HIV-1.
Zhou et al. screened for host factors that are required for HIV-1 replication using whole genome RNA interference (RNAi) technology (2008). Konig et al. also performed a global analysis of the host-pathogen interactions that regulate the early stages of HIV-1 replication (2008). In addition, other reports have also described cellular proteins interacting with HIV-1 proteins such as IN, Tat, and gp120 (Kashanchi et al., 1994; Wu et al., 1996; Ciuffi et al., 2005). Although a large number of interactions between HIV-1 proteins and host proteins have been identified, the information on binary interactions between HIV-1 viral proteins is relatively limited.
To increase our understanding of the role played by viral protein-protein interactions in the life cycle of HIV-1, we attempted to identify putative interactions among HIV-1 proteins. Using yeast two-hybrid assays, we identified seven interactions, including a novel interaction between gp41 and IN. The amino acids at positions 76–100 of gp41 were found to be responsible for the binding of gp41 to IN, as the deletion of this region within gp41 prevented its interaction with IN and reduced the production of HIV-1. This novel interaction may provide an insight into the life cycle of HIV-1.
-
For the yeast two-hybrid assays, each HIV-1 gene was amplified using PCR that used a plasmid carrying the WT HIV-1 strain, AD8, (pHIV-1-AD8) as a template. Each gene was cloned into both a pGADT7 vector (to be used as the bait vector) and a pGBKT7 vector (to be used as the prey vector), and they were designated pGADT7-p17, pGADT7-p24, pGADT7-NCp7, pGADT7-p6, pGADT7-IN, pGADT7-RT, pGADT7-PR, pGADT7-RH, pGADT7-Vif, pGADT7-Vpr, pGADT7-Vpu, pGADT7-Tat, pGADT7-Rev, pGADT7-gp120, pGADT7-gp41, pGADT7-Nef, pGBKT7-p17, pGBKT7-p24, pGBKT7-NCp7, pGBKT7-p6, pGBKT7-IN, pGBKT7-RT, pGBKT7-PR, pGBKT7-RH, pGBKT7-Vif, pGBKT7-Vpr, pGBKT7-Vpu, pGBKT7-Tat, pGBKT7-Rev, pGBKT7-gp120, pGBKT7-gp41, and pGBKT7-Nef. All the recombinant vectors were checked by carrying out restriction enzyme analyses and sequencing.
The yeast two-hybrid system employed the Saccharo-myces cerevisiae strain AH109 and the pGADT7 and pGBKT7 plasmids, along with plasmids that were used in the positive (the pair of pGADT7-T and pGBKT7-p53) and negative (the pair of pGADT7-T and pGBKT7-Lam) control assays. Each assay was carried out using the MatchmakerTM GAL4 Two-Hybrid System 3 (Clontech Laboratories, Palo Alto, USA) according to the manufacturer's instructions. Each recombinant vector was assayed on plates containing dropout supplements that lacked leucine, tryptophan, and histidine (-Leu/-Trp/-His), which allowed any potential autoactivation by the protein to be determined prior to the yeast two-hybrid assays being carried out.
The transformants that grew on the -Leu/-Trp/-His dropout medium were assessed for activation of the lacZ (the E. coli β-galactosidase gene involved in the hydrolysis of β-galactosides into monosaccharides) reporter gene using a filter lift assay according to the manufacturer's instructions (Clontech Laboratories). Protein-protein interactions were scored as positive when both the HIS3 (the yeast gene involved in the synthesis of histidine) and the lacZ reporter genes were activated.
In order to use the yeast two-hybrid system to map the region of gp41 that interacts with IN, cells of the yeast strain AH109 were transformed with gp41 deletion mutants (1–146, 147–191, 1–100, 101–191, 1–50, 51–100, 51–75, and 76–100) in a pGBKT7 plasmid and the pGADT7-IN plasmid, and they were assessed as described above.
-
To explore whether the peptide of aa 76–100 in gp41 is the region required for binding to IN. the pHIV-1-AD8Δgp41 (76–100) mutant plasmid was constructed. Briefly, two arbitrarily fragments, fragment 1 (nucleotides 6818–8026 of pHIV-1-AD8) and fragment 2 (nucleotides 8102–8848 of pHIV-1-AD8) were amplified using PCR with primer pairs A (sense: 5′-AAAAGGC-CTGTCCAAAGGTATCCTTTGAGC-3′; antisense: 5′-TAGTTGTCGATTTCTCTTTCCCACTCGCAGAT-GAGTTTTCCAGAGCAACCCCAAAT-3′) and B (sense: 5′-GATTTGGGGTTGCTCTGGAAAACTCATCTGC-GAGTGGGAAAGAGAAATCGACAACTAC-3′; antisense: 5′-CCGCTCGTGTCATTCTTTCCCTTACAGTA-G-3′) containing StuⅠ and BssSⅠ linker sequences (underlined), respectively. The two amplified DNA fragments were then subjected to overlap extension PCR to generate a single DNA fragment, followed by digestion with StuⅠ and BssSⅠ and cloning into a plasmid pHIV-1-AD8 that was digested with StuⅠ and BssSⅠ.
-
Vero (an African green monkey kidney cell line) and 293T (a human embryonic kidney cell line) cells were grown on Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum, 2 mmol/L Lglutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin (Life Technologies, Gaithersburg, USA) at 37 ℃ in 5% CO2. The cells were transfected with plasmids using LipofectamineTM 2000 Reagent (Life Technologies) according to the manufacturer's instructions. The transfected cells were cultured at 37 ℃ in 5% CO2 for 24–48 h for imaging and analysis.
-
The FRET analysis used an enhanced cyan fluorescent protein (ECFP) as the donor fluorophore coupled with an enhanced yellow fluorescent protein (EYFP) as the acceptor. The HIV-1-AD8 genes encoding IN and gp41 were amplified using PCR. Subsequently, the ORFs of the gp41 gene were inserted into a pECFP-C1 vector inframe with the ORF of ECFP using KpnⅠ and EcoR Ⅰ in order to construct the plasmid pECFP-gp41. The IN gene was inserted into a pEYFP-C1 vector in-frame with the ORF of EYFP using Hind Ⅲ and BamHⅠ in order to construct the plasmid pEYFP-IN. Vero cells were transfected with the two recombinant vectors as described above. At 24–30 h post-transfection, the cells were washed twice in phosphate-buffered saline (PBS, pH 7.2).
The FRET imaging and analysis was performed in Vero cells using a TCS SP2 Leica Laser Scanning Confocal Microscope (Leica, Heidelberg, Germany) equipped with a cooled charge-coupled device (CCD) camera. For live cell FRET imaging, the prepared cell culture dishes were placed in a temperature-controlled incubator at 37 ℃ and they were examined using a 63 × oil immersion objective with a numerical aperture of 1.32 and a 488 nm excitation beam. The excitation and emission bands for the cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) fluorescence channels used were as follows: CFP (left: 465 nm; right: 499 nm), YFP (left: 530 nm; right: 628 nm).
FRET measurements were performed using the FRET acceptor photobleaching procedure of the Leica confocal software, according to the manufacturer's instructions. The cells were bleached using a 514 nm laser beam at 100% intensity (95 mW of power at the specimen). We selected a region of interest (ROI) for photobleaching and the exposures lasted for 20–30 s.
Positive and negative controls were run in parallel with the experimental samples. The FRET efficiency (FRETeff) was calculated by the software according to the following equation: FRETeff=[Dpost–Dpre] / Dpost, where Dpost is the fluorescence intensity of the donor after acceptor photobleaching and Dpre is the fluorescence intensity of the donor before acceptor photobleaching. The statistical analysis was performed using SPSS 16.0 software (SPSS, Inc., Chicago, USA).
-
293T cells in 100 mm Petri dishes were transfected with 15 μg of pHIV-1-AD8 or pHIV-1-AD8Δgp41 (76–100). The total protein was harvested at 48 h post-transfection. Immunoprecipitation was performed using Dynabeads Protein G (Life Technologies) following the manufacturer's instructions. Briefly, an equal amount of beads was incubated with human monoclonal anti-gp41 antibody (IT-001-2F5, Immune Technology Corp, New York, USA) or mouse monoclonal anti-IN antibody (sc-69721, Santa Cruz Biotechnology, Inc., Santa Cruz, USA), or, in the case of the negative controls, with normal human IgG or normal mouse IgG. The same amount of total protein was then added to each beads-antibody complex. The immunoprecipitates were washed four times with cold PBS buffer and subjected to SDS-PAGE electrophoresis. The membranes were immunoblotted with either anti-gp41 or anti-IN primary antibody, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody.
-
A panel of gp41 deletion mutants was constructed and cloned into the plasmid pGBKT7 to test their interaction with pGADT7-IN by yeast two-hybrid assay. The interactions of gp41 deletion mutants with IN were quantified using an o-nitrophenyl-β-D-galactopyranoside (ONPG) assay to measure β-galactosidase activity, as described previously (Guarente, 1983; Mockli and Auerbach, 2004). Briefly, at least three independent clones were pelleted and resuspended in Z buffer, frozen in liquid nitrogen, and thawed at 37 ℃ in a water bath. Then the reaction systems (ONPG + Z buffer + β-mercaptoethanol + yeast cell resuspension) were placed in an incubator at 30 ℃. The reaction was stopped by adding 250 μL chilled 1 mol/L Na2CO3. The release of o-nitrophenol due to the hydrolysis of ONPG was assessed by measuring the absorbance at 420 nm and 600 nm. The β-galactosidase activity was expressed in arbitrary (Miller) units. The results were normalized to cell density and incubation time.
Yeast two-hybrid assay
Construction of pHIV-1-AD8Δgp41(76–100) mutant plasmid
Cell culture and transfection
FRET imaging and analysis
Co-IP assay and Western blotting
ONPG assay
-
To measure virus production, 293T cells seeded in 6-well plates were transfected with 0.25 μg of either pHIV-1-AD8 or pHIV-1-AD8Δgp41 (76–100) plasmids. After two days of culture, the supernatant was harvested and assessed for virus production using p24 ELISA, as described previously (Huang et al., 2007; Biswas et al., 2012). The samples were analyzed using a commercial ELISA kit (Beijing Key-Bio Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions.
-
To identify the protein-protein interactions between the HIV-1 proteins, we performed multiple yeast two-hybrid assays. The 15 viral proteins and the RNase H domain of the RT protein of HIV-1-AD8 were used. Thus a total of 16 proteins were tested, the main proposed functions of which are shown in Table 1.
Protein Function p17 Matrix protein; plasma membrane targeting of Gag for virion assembly; envelope incorporation; facilitates HIV-1 core into the cell nucleus; promotes proliferation and proinflammatory cytokines release p24 Capsid protein assembly into virion core structure p6 Vpr incorporation; virion budding p7 Encapsulates and protects viral genomic RNA; recognizes HIV-1 packaging signal of viral RNA RT RNA-dependent and DNA-dependent polymerase activity for cDNA synthesis RNase H Degrades RNA template strand from RNA/DNA duplex PR Proteolytic processing of Gag and Gag-pol polyproteins IN Catalyzes viral cDNA integration into the host cellular chromosome Rev Induces nuclear export of intron-containing viral RNAs; induces the transition from early to late phase of HIV-1 gene expression Tat Viral transcriptional transactivator; represses cellular promoters gp120 Interacts with CD4 receptor and chemokine co-receptors; mediates virus attachment and entry gp41 Non-covalently bound to gp120; mediates membrane fusion and virus entry Vpu CD4/MHC downregulation in endoplasmic reticulum; promotes virion release from host cell surface Vpr Promotes nuclear localization of preintegration complex; induces G2/M cell cycle arrest Vif Suppresses APOBEC3G/APOBEC3F; virion assembly Nef CD4/MHC downregulation; prevents apoptosis; increases efficiency of reverse transcription; T-cell activation Table 1. HIV-1 proteins analyzed in the yeast two-hybrid assays
Representative results for each of the protein-protein interactions identified in the yeast two-hybrid assay are shown in Figure 1. The assay identified seven interactions among HIV-1 proteins, six of which (gp120-tat, RH-RT, IN-RT, IN-RH, Rev-IN, and IN-Vpr) have been reported previously (Oz et al., 2002; Hehl et al., 2004; Marchio et al., 2005; Gleenberg et al., 2007; Rosenbluh et al., 2007; Levin et al., 2009). However, the gp41-IN interaction was a newly identified interaction and further experiments were performed to verify the finding.

Figure 1. Binary interactions between proteins of the HIV-1 strain AD8 that were screened using yeast twohybrid assays. Pairs of HIV-1 protein-encoding genes were inserted into a pGADT7 plasmid (to be used as the bait plasmid) or a pGBKT7 plasmid (to be used as the prey plasmid). The protein-protein interactions were screened after co-transformation of the bait and prey plasmids into AH109 yeast. Activation of the HIS3 reporter gene was assessed by assaying cell growth on media lacking histidine (His-). Activation of the lacZ reporter gene was assayed by staining for β-galactosidase activity. The photomicrographs demonstrate representative images obtained from three independent experiments. The asterisk indicates a novel interaction between gp41 and IN.
-
To confirm the gp41-IN interaction identified in the yeast two-hybrid assay, FRET imaging analyses were performed in live mammalian cells. We used an acceptor photobleaching method for the FRET analyses to detect the protein-protein interactions (Karpova et al., 2003). Specific areas of the cells were subjected to high strength illumination at the excitation wavelength of YFP (514 nm); in cases where there is a protein-protein interaction, this results in fluorescence depletion of the acceptor fluorophore (YFP) and an increased CFP signal.
As shown in Figure 2, selected ROIs of Vero cells that co-expressed ECFP-gp41 and EYFP-IN were bleached for 20–30 s using a 514 nm laser beam at 100% intensity and the fluorescence of ECFP (the donor fluorophore) and EYFP (the acceptor fluorophore) were imaged before and after photobleaching. The FRET images and the quantification analysis showed that an increase in the fluorescence intensity in the CFP channel could be detected with a concomitant decrease in YFP fluorescence. The FRETeff value for the assay of ECFP-gp41 and EYFP-IN was 8.49%±1.63% (n=14).
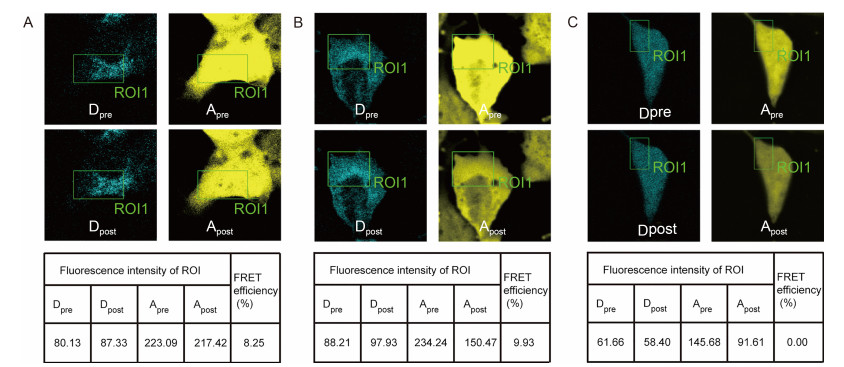
Figure 2. FRET images and analysis of the interactions between gp41 and IN in live cells. (A) A representative set of FRET images and quantitative analysis associated with the interaction between gp41-ECFP and IN-EYFP. During the acceptor photobleaching analysis, an increase in the fluorescence intensity in the ECFP channel was detected with a concomitant decrease in EYFP fluorescence, indicating an interaction between the proteins fused to ECFP and EYFP. (B) Results associated with the "cameleon" that was used as a positive control. (C) Results associated with the interaction between ECFP and EYFP, which was used as a negative control. ROI (region of interest) is the acceptor photobleaching area. Dpre and Apre are the fluorescence images of the donor and acceptor before acceptor photobleaching, respectively. Dpost and Apost are the fluorescence images of the donor and acceptor after acceptor photobleaching, respectively. The FRETeff value was calculated as FRETeff=[Dpost – Dpre] / Dpost. The photomicrographs demonstrate representative images obtained from three independent experiments.
In our experiments, the ECFP and EYFP pair was used as a negative control because they do not interact; accordingly, the negative control FRETeff value was close to zero. As a positive control, a plasmid encoding the tandem fusion protein "cameleon" was transfected into Vero cells for FRET analysis (Miyawaki et al., 1997). The FRETeff value of the positive control was 10.75%±1.42% (n=14). These negative and positive controls validated the method used in the FRET analyses for the detection of protein-protein interactions. The FRET analysis indicated the presence of gp41-ECFP and IN-EYFP interactions in live cells and thereby verified the newly identified gp41-IN interaction.
-
Both the yeast two-hybrid assays and live cell FRET analyses that involved transfected cells identified the interaction between gp41 and IN. However, these assays were carried out in the absence of other viral factors and so the results may not necessarily have reflected the interactions that occur during the course of virus replication.
In 293T cells transfected with pHIV-1-AD8 that encoded AD8 proviral cDNA, HIV-1 proteins were expressed and progeny virions could be produced, mimicking HIV-1 replication. To assess whether the newly identified gp41-IN interaction could be demonstrated in these circumstances, cell lysate was prepared from the 293T cells transfected with pHIV-1-AD8 at 48 h posttransfection and immunoprecipitation was carried out using specific antibodies.
Subsequently, the samples were subjected to SDSPAGE and immunoblotting to establish whether the interacting proteins were present. IN was found to coprecipitate with gp41 (Figure 3A). The reciprocal Co-IP experiment also showed that gp41 could be found in the IN precipitate (Figure 3B). No corresponding immunoreactive bands were detected when normal IgG was used in the assay. These results confirmed that IN indeed interacts with gp41 in the context of pHIV-1-AD8 transfected cells.
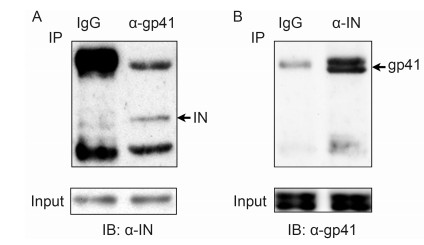
Figure 3. Interaction of gp41 and IN confirmed using Co-IP assays. (A) Co-IP assay using anti-gp41 antibody and extracts of 293T cells that had been transfected with pHIV-1-AD8. The Western blot was carried out with anti-IN antibody. (B) Reciprocal Co-IP assay using anti-IN antibody, which confirmed the interaction between gp41 and IN. The Western blot was carried out with antigp41 antibody. The photomicrographs demonstrate representative images obtained from three independent experiments.
-
To determine the region of gp41 that is essential for binding to IN, a panel of gp41 deletion mutants were each incorporated into a pGBKT7 plasmid and these plasmids were each individually introduced into yeast cells along with a pGADT7-IN plasmid. As shown in Figure 4, initially, gp41 was split in half into N-terminal and C-terminal fragments and the former fragment was found to interact with IN. Subsequently, the IN-interacting fragment of gp41 was further serially split in half and the interactions of the resultant fragments with IN were assessed. β-galactosidase activity was assayed and the amino acids at positions 76–100 in gp41 were found to be associated with its capacity to bind to IN.
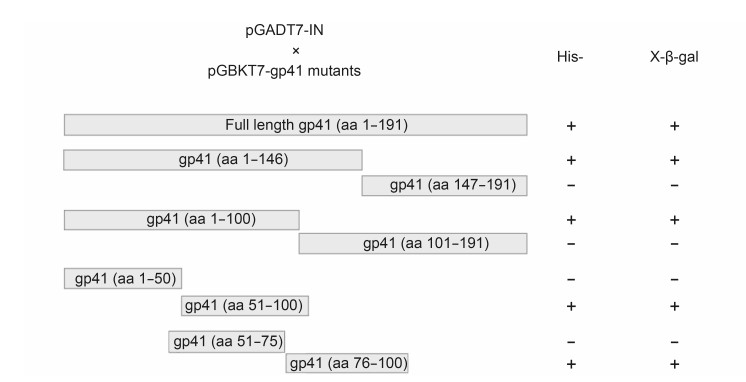
Figure 4. Identification of the amino acid sequence of gp41 associated with its capacity to bind to IN using serial deletion constructs of gp41. A panel of gp41 deletion mutants was constructed and each was cloned into a pGBKT7 plasmid to test the interaction with pGADT7-IN using yeast two-hybrid assays. "+" and "?" indicate growth and no growth of AH109 yeast transformants on media lacking histidine (His-), or staining and no staining associated with β-galactosidase activity. One representative result of three independent experiments is shown.
Additionally, β-galactosidase assays were performed in order to quantify the strength of the interaction of individual gp41 deletion mutants with IN (Figure 5). The interaction between gp41 and IN was weaker than that between p53 and large T antigen, which were used as a positive control. However, the ONPG assays gave comparable results regarding the gp41 and IN interaction and the RT and IN interaction, which had already been validated in previous studies (Wilkinson et al., 2009; Tekeste et al., 2015).
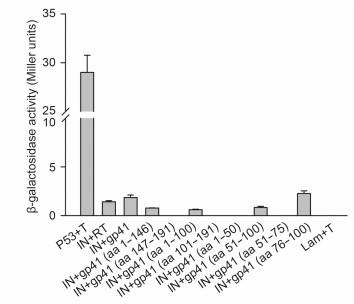
Figure 5. Quantitative analysis of protein-protein interactions as determined using an o-nitrophenyl-β-Dgalactopyranoside (ONPG) assay. Transformants of p53/T, Lam/T, and IN/RT were used as controls. The β-galactosidase activity in each transformant was monitored using a liquid ONPG assay and the results are shown in Miller units. The error bars show the means±the standard deviations of three independent experiments.
In summary, the gp41 deletion mutants containing amino acids at positions 1–146, 1–100, 51–100, and 76–100 of gp41 retained β-galactosidase activity, while the constructs containing amino acids at positions 147–191, 101–191, 1–50, and 51–75 had no β-galactosidase activity. Taken together, the region containing amino acids at positions 76–100 of gp41 appeared to be the region required for binding to IN.
-
To further confirm that the gp41 (76–100) region is responsible for the gp41-IN interaction, a pHIV-1-AD8Δgp41 (76–100) mutant plasmid was generated. The construct encoding this gp41 deletion mutant was introduced into 293T cells, and the effect of the deletion on the gp41-IN interaction was determined using a Co-IP assay. As expected, deletion of gp41 (76–100) resulted in the production of a truncated gp41 protein (of approximately 36 kDa), which was recognized by the anti-gp41 antibody. The deletion of the gp41 (76–100) region was shown to almost completely prevent gp41 from binding to IN in the Co-IP assay (Figure 6A and 6B). This suggested that though the gp41 (76–100) region is not essential for the expression of gp41, it is required for gp41 to bind to IN.
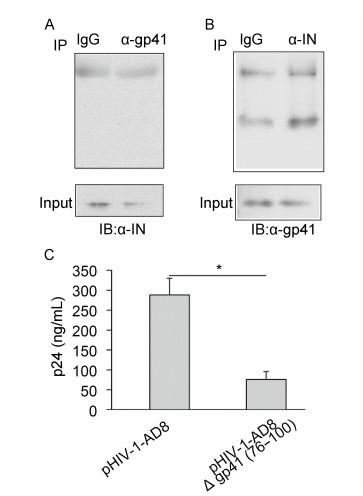
Figure 6. Results of Co-IP assays and p24 ELISA indicating that the deletion of the gp41(76–100) region prevented the gp41-IN interaction and reduced virus production. (A) Co-IP assay using anti-gp41 antibody and extracts of 293T cells that had been transfected with pHIV-1-AD8Δgp41(76–100). The Western blot was carried out with anti-IN antibody. (B) Co-IP assay using anti-IN antibody and extracts of 293T cells that had been transfected with pHIV-1-AD8Δgp41(76–100). The Western blot was carried out with anti-gp41 antibody. The photomicrographs demonstrate representative images obtained from three independent experiments. (C) HIV-1 p24 levels in the supernatants of cell cultures of 293T cells that had been transfected with either pHIV-1-AD8 or pHIV-1-AD8Δgp41(76–100) plasmids. The error bars show the means±the standard deviations of three independent experiments. The asterisk indicates a statistically significant difference (P < 0.05) between the two groups according to Student's t-test.
To assess the effect of this mutant in the context of virus replication, both a pHIV-1-AD8 plasmid and a pHIV-1-AD8Δgp41 (76–100) plasmid were separately transfected into 293T cells. We examined the effect of the mutant on the production of the HIV-1 p24 capsid protein using the culture supernatants and a p24 ELISA. In the cells that were transfected with pHIV-1-AD8Δgp41 (76–100) there was evidence of marked decreased in the levels of p24 compared with the levels in the cells that were transfected with pHIV-1-AD8 (Figure 6C). Thus, deletion of the gp41 (76–100) region was shown to reduce virus production.
Identification of protein-protein interactions using yeast two-hybrid assays
Live cell FRET imaging and analysis of the gp41-IN interaction
Co-IP assay of the gp41-IN interaction.
Identification of gp41 sequence responsible for its interaction with IN
Effect of the deletion of gp41(76–100) on the gp41-IN interaction and virus production
-
Viral protein-protein interactions are pivotal for many of the functions of the viral proteins and they play critical roles in multiple processes in the virus life cycle. In this study we screened for binary interactions between HIV-1 proteins using yeast two-hybrid assays. Seven proteinprotein interactions were identified in our experiments. Of these interactions, six (gp120-tat, RH-RT, Rev-IN, IN-Vpr, IN-RT, and IN-RH) have been previously reported in the literature. These protein-protein interactions have been shown to be important for the infection of cells by HIV-1. For example, the IN-RT interaction stimulates the IN-mediated strand transfer reaction (Oz et al., 2002; Hehl et al., 2004). Furthermore, Rev and Vpr can interact with IN and inhibit its activity (Rosenbluh et al., 2007; Levin et al., 2009).
Our yeast two-hybrid assays also revealed a novel interaction between gp41 and IN, which was subsequently confirmed by both the FRET analysis and the Co-IP assays. However, several previously reported interactions between HIV-1 proteins were not detected in the present study, including gp41-p17, gp41-gp120, Vpr-p6, and Vpr-NCp7 interactions (de Rocquigny et al., 1997; Wyatt et al., 1997; Selig et al., 1999; Murakami and Freed, 2000). It could be postulated that several factors, such as differences in protein folding, the absence of posttranslational modifications, and insufficient sensitivity, may have led to false negatives in our yeast two-hybrid assays.
The interaction between gp41 and IN is an interesting finding. It is known that gp41 mediates membrane fusion and virus entry (Chan et al., 1997) whereas IN mediates the insertion of the viral DNA into the host chromosomal DNA (Chen et al., 2000). Even though gp41 and IN have been found to be located separately in mature HIV-1 virions, it is possible that the gp41-IN interaction could occur in the context of the virus replication cycle. It has been shown that Env-derived peptides in gp41 can cause a remarkable inhibition of the strand transfer and 3′-end processing reactions that are catalyzed by IN (Suzuki et al., 2010). This suggests that gp41 may affect the activity of IN via protein-protein interaction. Several studies of interactions between HIV-1 structural and non-structural proteins have been reported, such as the gp120-tat, (Marchio et al., 2005), Vpr-p6 (Jenkins et al., 2001), and Vpr-NCp7 interactions (de rocquigny et al., 1997). The gp41-IN interaction identified in this study provides further evidence of interactions between the structural and non-structural gene products of HIV-1, which may have functions in the viral life cycle.
In this study, the amino acids at positions 76–100 of gp41 were found to be required for binding to IN. HIV-1 gp41 consists of an extracellular domain (an ectodomain), a transmembrane domain, and an intracytoplasmic tail domain. The ectodomain contains a hydrophobic N-terminal fusion peptide, an N-terminal heptad repeat (NHR), a disulfide-bridged loop region, a C-terminal heptad repeat (CHR), and a membrane-proximal external region (Roche et al., 2014). The gp41 (76–100) region is contained within the loop region, which is ordered and displays numerous intermolecular and nonsequential intramolecular contacts. For instance, the gp41 loop region has been shown to interact with the C1 and C5 regions of gp120 and C1q, a component of the host complement system (Caffrey et al., 1998; Caffrey, 2001).
To gain an insight into the possible functional connections between gp41 and IN, a mutant HIV-1 plasmid with a deletion in the gp41 (76–100) region was constructed and the subsequent Co-IP assays indicated that the deletion prevented the gp41-IN interaction. Deletion of the gp41 (76–100) region was also associated with reduced virus production. These results demonstrate that the gp41 (76–100) region is crucial for the interaction between gp41 and IN, reinforcing the findings regarding the role of the gp41 loop region in mediating intermolecular contacts.
HIV-1 production involves a highly orchestrated series of interactions between proteins encoded by the virus, and between viral proteins and key cellular components (Freed, 2004; Goff, 2007). For instance, a fragment of integrase interactor 1 (INI1, a protein that interacts with HIV-1 IN) potently inhibits HIV-1 production. Disruption of the IN-INI1 interaction prevents the inhibitory effect (Yung et al., 2001). These findings indicate that IN may play a role in the process of virus production in addition to its role in integration (Shin et al., 1994; Bukovsky and Gottlinger, 1996). Although the basis for the impact of the gp41-IN interaction on virus production remains to be elucidated, it is likely that interference with this interaction affects the virus production process. Further investigation is warranted to unravel the detailed molecular mechanisms underlying the role of the gp41-IN interaction in the HIV-1 life cycle.
In conclusion, we explored the binary interactions between HIV-1 proteins and discovered a new interaction between gp41 and IN. In addition, we determined that the amino acids at positions 76–100 of gp41 are required for it to bind to IN. Deletion of this region within gp41 prevented its interaction with IN and reduced HIV-1 production. This newly identified protein-protein interaction is likely to be an important factor in the life cycle and pathogenesis of HIV-1.
-
We thank Yuntao Wu from George Mason University for providing the pHIV-1-AD8 plasmid. We are grateful to Yizhou Gao for his contribution to the construction of the plasmids and the yeast-two hybrid assays. We also thank the Core Facility and Technical Support at the Wuhan Institute of Virology for their excellent technical support. ZQ Cui was supported by the National Nano Project (grant no. 2011CB933600) and the National Natural Science Foundation of China (grant no. 31470269). XE Zhang was supported by the Institute of Biophysics and the National Laboratory of Biomacromolecules at the Chinese Academy of Sciences (CAS). The authors are grateful for the support they received from the CAS.
-
The authors declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
-
ZC and XEZ conceived of the study and designed the experiments. XWZ, FZ, XM and XZ participated in yeast two-hybrid screen assay. XWZ, FZ and XM performed Co-IP and WB experiments. WL, ZZ and JZ carried out the FRET assay. ZC and XWZ drafted the manuscript. All authors read and approved the final manuscript.














 DownLoad:
DownLoad: