-
Dear Editor,
In this study, we use SRLCA to identify the possible Hsp90-ORF36 interaction. Additionally, critical amino acids of ORF36 and Hsp90 are deleted or mutated to evaluate their possible contributions to Hsp90-ORF36 interaction. The results indicated that the aa 215–257 of ORF36 and aa 110–180 region of Hsp90 are essential for the Hsp90-ORF36 interaction. The M216, L221, D225, F226, W237, and T238 in ORF36 and A121, Q133, F134, T149, E158, and G168 in Hsp90 are important for the Hsp90-ORF36 interaction. SRLCA is based on the complementation of the N-terminal domains of Renilla luciferase (LN) and C-terminal domains of Renilla luciferase (LC) non-functional halves of Renilla luciferase fused to possibly interacting proteins and emit luminescence (Deng et al., 2011; Jiang et al., 2010)(Supplementary Figure S1A). The Kaposi's sarcoma-associated herpesvirus open reading frame 36 (ORF36) encodes a serine protein kinase, playing an important role in the development of malignancy (Kim et al., 2013). Heat shock protein 90 (Hsp90) plays crucial roles in post-translational folding and stability of many proteins (Chehab et al., 2015; Pennisi et al., 2015). In this study, we use SRLCA to identify the possible Hsp90-ORF36 interaction. Additionally, critical amino acids of ORF36 and Hsp90 are deleted or mutated to evaluate their possible contributions to Hsp90-ORF36 interaction. The results indicated that the aa 215–257 of ORF36 and aa 110–180 region of Hsp90 are essential for the Hsp90-ORF36 interaction. The M216, L221, D225, F226, W237, and T238 in ORF36 and A121, Q133, F134, T149, E158, and G168 in Hsp90 are important for the Hsp90-ORF36 interaction.
The plasmids pcDNA3.1, PA-LN and PA-LC were obtained from Prof. Feng Li (South Dakota State University, South Dakota State, USA). The first group of full length KSHV open reading frame 36 (ORF36) and heat shock protein 90 (Hsp90) sequences were PCR-amplified using the primers designed with Xho I and EcoR I (NEB, USA), and subcloned upstream of LN and LC with a linker DNA sequences GGGGSGGGGS ((G4S)2) (Paulmurugan and Gambhir, 2003). ORF36 plasmids were tagged by Flag. The second group of full-length KSHV ORF36 and Hsp90 sequences were PCR-amplified using the primers designed with Xho I and NotI, and subcloned downstream of LN and LC with a linker (G4S)2 (Supplementary Figure S1B, S1C). All plasmids were detected in HEK293 cells (Supplementary Figure S2A). All possible combinations of Hsp90-ORF36pairs were transfected into 293T cells for 48 h and detected the luciferase activity. As shown in Figure 1B, the pair of Hsp90-LC+ORF36-LN has the highest luciferase activity(~22827 RLU). These results suggest that the pair Hsp90-LC/ORF36-LNhas the most powerful interaction. Therefore, thepair of Hsp90-LC/ORF36-LNwas selected to study the Hsp90-ORF36interactions through the SRLCA. Additionally, according to our previous study (Wang et al., 2016), deletion mutants of ORF36 were constructed to confirm the essential amino acids region. Most of the polar amino acids in this region were mutated. Moreover, several non-polar amino acids were randomly selected to mutate in this region. All deletion and point mutation of ORF36 were linked to upstream of LN. The recombinant plasmids ORF36d(1-145)-LN and ORF36d(290-445)-LN were PCR-amplified from ORF36-LN. Primers were designed by Primer5 as follows: ORF36d(1-145) Xho I-F/ORF36 EcoR I-R, ORF36 Xho I-F Flag/ORF36d(290-445)-R. The plasmid ORF36d(140-295)-LN was amplified from ORF36-LN. Primers were designed by Primer5 as follows: ORF36 Xho I-F Flag/ORF36d(140-295)-R, ORF36d(140-295)-F/ORF36 EcoR I-R Flag. After the first PCR-amplification, the products were mixed and amplified for ten cycles using primers ORF36 Xho I-F Flag/ORF36 EcoR I-R Flag. The plasmids ORF36d(130-183)-LN, ORF36d(172-226)-LN, ORF36d(215-257)-LN, and ORF36d(247-308)-LN were constructed by the same method. The mutants ORF36(R220A)-LN, ORF36(S228A)-LN, ORF36(M216A)-LN, ORF36(L221A)-LN, ORF36(D225A)-LN, ORF36(F226A)-LN, ORF36(W237A)-LN, and ORF36 (T238A)-LN were amplified from ORF36-LN. The primers were designed by Primer 5 as follows: ORF36(R220A)-F/R, ORF36(S228A)-F/R, ORF36(M216A)-F/R, ORF36(L221A)-F/R, ORF36(D225A)-F/R, ORF36(F226A)-F/R, ORF36(W237A)-F/R, and ORF36(T238A)-F/R. All deletion and point mutations of Hsp90 were linked to upstream of LC by the same way. All primers used in this study were showed in Supplementary Figure S1.
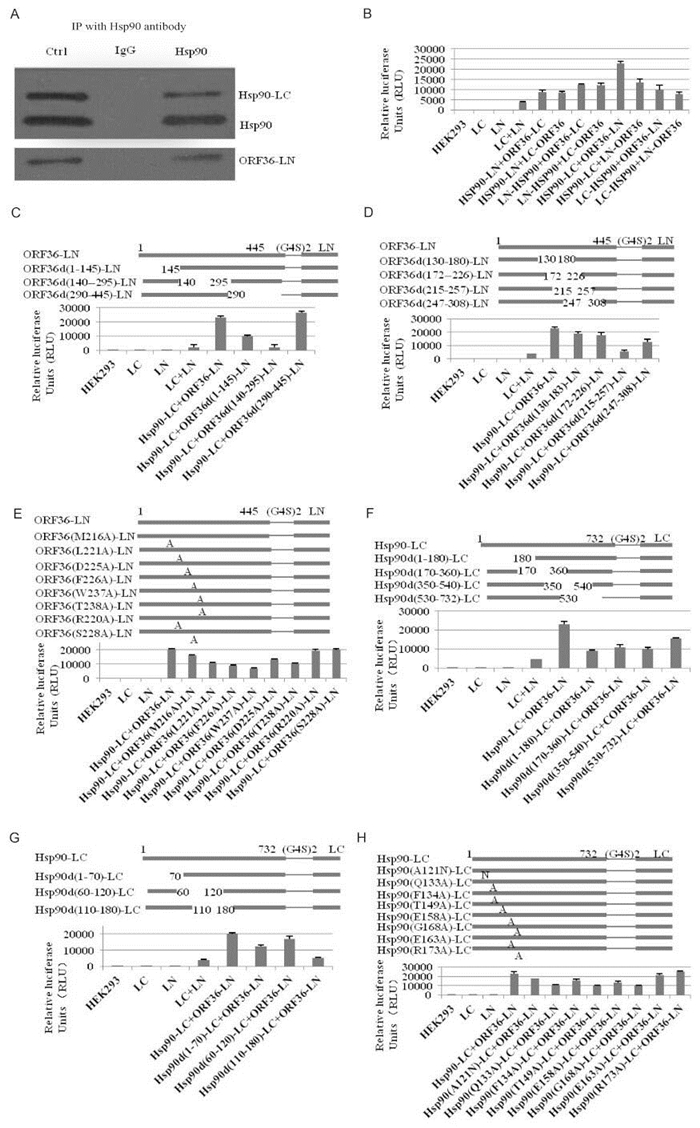
Figure 1. PPIs was detected by IP and Luciferase assay. (A)The plasmids Hsp90-LC and ORF36-LN were transiently co-transfected into HEK293 cells. Cell lysates were immunoprecipitated with Hsp90 antibody. IgG was used as a negative control. Cell lysates without immunoprecipitation were used as control.(B) HEK293 cells were transfected with various plasmids. Luciferase assay was performed under experimental procedures. Data are presented as mean ± SD (n=6). (C, D and E) The recombinant mutants of ORF36 linked to LN via the linker (G4S)2. Luciferase assay of HEK293cells transfected with these mutants after 48 h. Data are presented as mean ± S.D. (n =6). (F, G and H) The recombinant mutants of Hsp90 linked to the LC via the linker (G4S)2. Luciferas e assay was performed under experimental procedures. Data are presented as mean ± SD (n=6).
To determine if the expressed fusion proteins (Hsp90-LC and ORF36-LN) can interact with each other, IP was performed to detect Hsp90-ORF36 interaction after transfection.As shown in Figure 1A, both wild-type Hsp90 and fusion protein Hsp90-LC were precipitated by Hsp90 antibody, and ORF36-LN could be detected by Flag antibody. These results suggest that the fusion proteins (Hsp90-LC and ORF36-LN) are able to interact with each other in living cells.
To detect complementation of Hsp90-LC and ORF36-LN, HEK293 cells were transfected with various constructs as indicated. As shown in Figure 1B, when the HEK293 cells were transfected with each fragment alone (LN, LC, LN+LC), the luciferase activity was low. However, when the two plasmids Hsp90-LC and ORF36-LN were co-transfected into HEK293 cells, the luciferase activity was elevated (about 22827 RLU). These results suggest that the fusion proteins (Hsp90-LC and ORF36-LN) are able to interact with each other in cells.
To determine the essential region of ORF36 for the Hsp90-ORF36 interaction, deletion mutants of ORF36 (ORF36d(1-145)-LN, ORF36d(140-295)-LN, ORF36d(290-445)-LN) were constructed. All mutants can be detected in HEK293 cells (Supplementary Figure S2B). As demonstrated in Figure 1C, co-transfection of Hsp90-LC/ORF36d(140-295)-LN only restored 9.05% luciferase activity compared with full length Hsp90-LC/ORF36-LN. However, co-transfection of Hsp90-LC/ORF36d(1-145)-LN and Hsp90-LC/ORF36d(290-445)-LN restored 41.67% and 105.53% luciferase activity compared with full length Hsp90-LC/ORF36-LN. These results suggest thatthe aa140-295 region of ORF36 is essential for the Hsp90-ORF36 interaction.Therefore, four recombinant plasmids (ORF36d(130-183)-LN, ORF36d(172-226)-LN, ORF36d(215-257)-LN, and ORF36d(247-308)-LN) were constructed from the wild type of ORF36-LN.The results of SRLCA indicated that deletion of the aa215-257 region in ORF36 only restored 22.90% luciferase activity compared with full-length of Hsp90-LC/ORF36-LN. However, the luciferase activity of Hsp90-LC/ORF36-LN was not decreased significantly by Hsp90-LC/ORF36d(130-183)-LN, Hsp90-LC/ORF36d(172-226)-LN, and Hsp90-LC/ORF36d(247-308)-LN (Figure 1D). These results suggest the aa215-257 region of ORF36 is crucial in the ORF36-Hsp90 interaction.
Since the above results indicated that the aa215-257 region of ORF36 is important for Hsp90-ORF36 interaction, most of the polar amino acids in this region were mutated. Moreover, several non-polar amino acids were randomly selected to mutate in this region. Here we show the most important amino acids M216, L221, D225, F226, W237, and T238 of ORF36. In addition, R220 and S228 were used as the negative controls. All mutants can be detected in HEK293 cells (Supplementary Figure S2D). The result of SRLCA showed us that luciferase activity were not changed by Hsp90-LC/ORF36(R220A)-LN or Hsp90-LC/ORF36(S228A)-LN. The mutations in ORF36 (M216A, L221A, D225A, F226A, W237A, and T238A) reduced the luciferase activity by 22.55%, 47.97%, 35.10%, 57.19%, 69.44%, and 49.80%, respectively, compared with the full-length Hsp90-LC/ORF36-LN (Figure 1E). The data indicate that M216, L221, D225, F226, W237, and T238in ORF36 are essential for Hsp90-ORF36 interaction.
To investigate the functional region of Hsp90 for the Hsp90-ORF36 interaction, deletion mutants of Hsp90 (Hsp90d (1-180)-LC, Hsp90d(170-360)-LC, Hsp90d(350-540)-LC, and Hsp90d (530-732)-LC) were constructed. These mutants of Hsp90-LC can be detected by Hsp90 antibody (Supplementary Figure S2E). The result of SRLCA showed us that Hsp90d(1-180)-LC/ORF36-LN restored about 39.07% luciferase activity compared with full-length Hsp90-LC/ORF36-LN, which was lower than others (Figure 1F). Therefore, three recombinant plasmids (Hsp90d(1-70)-LC, Hsp90d(60-120)-LC, Hsp90d(110-180)-LC) were constructed. These mutants can be detected by Hsp90 antibody (Supplementary Figure S2F). The result of SRLCA showed us that deletion of the aa 110-180 region in Hsp90 reduced Hsp90-ORF36 interaction by 72.73% compared with wild type of Hsp90-LC and ORF36-LN (Figure 1G). The data suggest that the aa 110-180 region of Hsp90 is essential for the Hsp90-ORF36 interaction.
To investigate the important amino acids of Hsp90 for the Hsp90-ORF36 interaction, most of the polar amino acids in this region (110-180) were mutated. Moreover, several non-polar amino acids were randomly selected to mutate in this region. Here we show the most important amino acids A121, Q133, F134, T149, E158, and G168 of Hsp90 were mutated. In addition, E163 and R173 were used as the negative controls. All point mutants of Hsp90-LC can be detected by Hsp90 antibody (Supplementary Figure S2G). The result of SRLCA indicated that the Hsp90-LC/ORF36-LN interaction was not changed by Hsp90(E163A)-LC/ORF36-LN or Hsp90(R173A)-LC/ORF36-LN. However, Hsp90(A121N)-LC/ORF36-LN, Hsp90(Q133A)-LC/ORF36-LN, Hsp90(F134A)-LC/ORF36-LN, Hsp90(T149A)-LC/ORF36-LN, Hsp90(E158A)-LC/ORF36-LN, and Hsp90(G168A)-LC/ORF36-LN abolished Hsp90-ORF36 interaction by 75.76%, 46.17%, 68.26%, 42.64%, 50.02%, and 41.25%, respectively, as measured by the restored luciferase activity (Figure 1H). These results suggest that A121, Q133, F134, T149, E158, and G168in Hsp90 are directly involved in the Hsp90-ORF36 interaction.
PPIs are believed to play complex roles in many cellular process, such as transcription, translation, signal transduction, cell division, and oncogenic transformation (Zhang et al., 2016). The inhibition of Hsp90 has become a new strategy for cancer therapeutics (Peng et al., 2007). The functional Hsp90 requires cochaperones to form the Hsp90 chaperone machinery, which plays a key role in orchestrating the spatial and temporal order of protein interactions (Pennisi et al., 2015). Therefore, investigation of PPIs roles in Hsp90 superchaperone machinery may offer a new therapeutic strategy for virus infections and the associated diseases.
In this study, SRLCA was used to study full-length Hsp90-ORF36 interaction in living cells. Furthermore, we evaluated the contributions of the essential amino acids in the Hsp90-ORF36 interaction. SRLCA can be used alone or associated with IP or other techniques to detect PPIs.
In summary, the Hsp90-ORF36 interaction was tested using SRLCA method, and the interacting motifs were investigated. It is found that the aa215-257 of ORF36 and aa110-180 region of Hsp90 are essential for the Hsp90-ORF36 interaction. The M216, L221, D225, F226, W237, and T238 in ORF36 and A121, Q133, F134, T149, E158, and G168 in Hsp90 are important for the Hsp90-ORF36 interaction. These results provide a rationale to develop inhibitors for disruption of the Hsp90-ORF36interactions.
HTML
-
The study was supported by the Initiative Research Program of Wuhan University (No.410100020), the advanced talent independent research program of Wuhan University (No. 410100011) and the National Natural Science Foundation of China (No. 210700228). The authors declare no conflicts of interest. This article does not contain any studies with human or animal subjects performed by any of the authors.
Supplementary figures/table are available on the websites of Virologica Sinica:www.virosin.org; link.springer.com/journal/12250
-
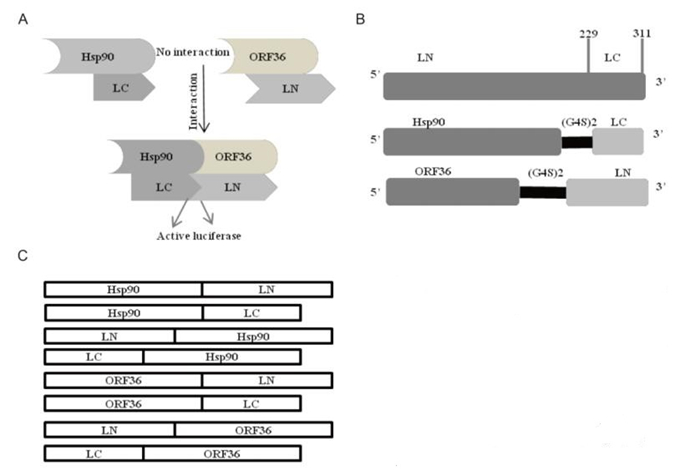
Figure S1. Schematic diagram of SRLCA. (A) The interaction between Hsp90 and ORF36 bring LN and LC closely enough and lead to complementation of Renilla luciferase enzyme activity and emit luminescence. (B) Renilla luciferase is an enzyme including LN (aa1-229) and LC (aa230-311) fragments. ORF36 and Hsp90 are fused to the LN and LC domains through a linker (G4S)2. (C) Schematic of plasmids used in this study.
Figure S2. Expression of plasmids was confirmed by western blotting. (A) The recombinant plasmids Hsp90-LC, LC-Hsp90, Hsp90-LN, LN-Hsp90, ORF36-LC, LC-ORF36, ORF36-LN, and LN-ORF36 were transfected into HEK293.After 48 h, cell lysates were collected and subjected to western blotting. (B, C and D) The recombinant mutants of ORF36 were transfected into HEK293 cells. After 48 h, cell lysates were collected and subjected to western blotting. (E, F, and G) The recombinant mutants of Hsp90 were transfected into HEK293 cells. After 48 h, cell lysates were collected and subjected to western blotting.
primer name primer sequence (5'-3') ORF36 Xho I-F Flag ATCGCTCGAGATGGATTACAAGGATGACGACGATAAGATGCGCTGGAAGAGAATGGAGAGG ORF36 EcoR I-R Flag TAGCGAATTCGCGAAAACAAGTCCGCGGGTGTGG Hsp90 Xho I-F ATCGCTCGAGATGCCTGAGGAAACCCAGACCCAAG Hsp90 EcoR I-R ATCGGAATTCCTTGGGTCTGGGTTTCCTCAGGCAT ORF36d(1-145)-F TACGCTCGAGATGGATTACAAGGATGACGACGATAAGATGGTGCAGCCTTGCATACCCTG ORF36d(1-145)-R TAGCGAATTCGCGAAAACAAGTCCGCGGGTGTGG ORF36d(140-295)-F CTGCTAGGATTCCCTAACATGGGACTG ORF36d(140-295)-R CAG TCC CATGTTAGG GAATCCTAGCAG ORF36d(290-445)-F ATCGCTCGAGATGGATTACAAGGATGACGACGATAAGATGCGCTGGAAGAGAATGGAGAGG ORF36d(290-445)-R GTACGAATTCGCTACCTGCGCGTGCATCTTACCG ORF36d(130-183)-F CGAGAGAGGTACCCCGCGGCGGAGTTC ORF36d(130-183)-R GAACTCCGCCGCGGGGTACCTCTCTCG ORF36d(172-226)-F CAGGTCAACCTGAGGCTGGTCCTTA ORF36d(172-226)-R TAAGGACCAGCCTCAGGTTGACCTG ORF36d(215-257)-F CGCCCATGAACCCCTACGACTT ORF36d(215-257)-R AAGTCGTAGGGGTTCATGGGCG ORF36d(247-308)-F CTGGGGTTTAAGCAAATCGACATGTCC ORF36d(247-308)-R GGACATGTCGATTTGCTTAAACCCCAG ORF36(R220A)-F CCCATGAACCCCGGGGCCCTGGTCCTT ORF36(R220A)-R AAGGACCAGGGCCCCGGGGTTCATGGG ORF36(S228A)-F ACCGATTTCGGTGCAGTTGCGCTA ORF36(S228A)-R TAGCGCAACTGCACCGAAATCGGT ORF36(M216A)-F GACCTAACGCCCGCTAACCCC ORF36(M216A)-R GGGGTTAGCGGGCGTTAGGTC ORF36(L221A)-F ATGAACCCCGGGAGGGCTGTCCTTACCGATTTC ORF36(L221A)-R CGGGGACAGCCCTCCCGGGGTTCAT ORF36(D225A)-F GTCCTTACCGCTTTCGGTTCCGTTGCG ORF36(D225A)-R CAACGGAACCGAAAGCGGTAAGGAC ORF36(F226A)-F GTCCTTACCGATGCTGGTTCCGTTGCG ORF36(F226A)-R CAACGGAACCAGCATCGGTAAGGAC ORF36(W237A)-F CACTCTGGGAGCAAGGCTACTAACCTT ORF36(W237A)-R GGTTAGTAGCCTTGCTCCCAGAGTG ORF36(T238A)-F GGGAGCAAGTGGGCTAACCTTGTGGTGACC ORF36(T238A)-R CCACAAGGTTAGCCCACTTGCTCCC Hsp90d(1-180)-F ATGCCTCGAGGGTCGTGGAACAAAAGTTATCCTAC Hsp90d(1-180)-R ATCGGAATTCCTTGGGTCTGGGTTTCCTCAGGCAT Hsp90d(170-360)-F TTCACAGTGAGGACAAAAAAGAACAAT Hsp90d(170-360)-R ATTGTTCTTTTTTGTCCTCACTGTGAA Hsp90d(350-540)-F GATCTGTTTGAAAACTTTGAGGGGAAG Hsp90d(350-540)-R CTTCCCCTCAAAGTTTTCAAACAGATC Hsp90d(530-732)-F GTCCAACAGCTGAAGATGGAAGAAGTA Hsp90d(530-732)-R TACTTCTTCCATCTTCAGCTGTTGGAC Hsp90d(1-170)-F ATGCCTCGAGGACTCTGGGAAAGAGCTGCATATTA Hsp90d(1-70)-R ATCGGAATTCCTTGGGTCTGGGTTTCCTCAGGCAT Hsp90d(60-120)-F CGGTATGAAAGCTTGAAAGCGTTCATG Hsp90d(60-120)-R CATGAACGCTTTCAAGCTTTCATACCG Hsp90d(110-180)-F ATCGCCAAGTCTGGGACAGGTGAACCT Hsp90d(110-180)-R AGGTTCACCTGTCCCAGACTTGGCGAT Hsp90(A121N)-F GCGTTCATGGAAAACTTGCAGGCT Hsp90(A121N)-R AGCCTGCAAGTTTTCCATGAACGC Hsp90(Q133A)-F TCTATGATTGGCGCGTTCGGT Hsp90(Q133A)-R ACCGAACGCGCCAATCATAGA Hsp90(F134A)-F ATGATTGGCCAGGCGGGTGTTGGT Hsp90(F134A)-R ACCAACACCCGCCTGGCCAATCAT Hsp90(T149A)-F GCTGAGAAAGTAGCAGTGATCACCAAA Hsp90(T149A)-R TTTGGTGATCACTGCTACTTTCTCAGC Hsp90(E158A)-F CATAACGATGATGCACAGTACGCTTGG Hsp90(E158A)-R CCAAGCGTACTGTGCATCATCGTTATG Hsp90(G168A)-F GAGTCCTCAGCAGGGGCATCATTCACAGTG Hsp90(G168A)-R CACTGTGAATGATGCCCCTGCTGAGGACTC Hsp90(E163A)-F CAGTACGCTTGGGCATCCTCAGCAGGG Hsp90(E163A)-R CCCTGCTGAGGATGCCCAAGCGTACTG Hsp90(R173A)-F TCATTCACAGTGGCCACAGACACAGGT Hsp90(R173A)-R ACCTGTGTCTGTGGCCACTGTGAATGA Table 1. Primers used in this study.
















 DownLoad:
DownLoad: