HTML
-
Hantaviruses are enveloped single-stranded RNA viruses belonging to the Bunyaviridae family, Hantavirus genus, and are mainly transmitted to humans through inhalation of contaminated aerosols of rodent excreta. Hantavirus infections can lead to two severe diseases in humans worldwide, i.e., hemorrhagic fever with renal syndrome (HFRS) in Eurasia, caused by Old World hantaviruses, and hantavirus pulmonary syndrome (HPS) in North and South America, caused by New World hantaviruses. The global incidence of HFRS and HPS in humans exceeds 150, 000 cases per year. China is a highly endemic HFRS area and accounts for 90% of the total cases worldwide. Hantaan virus (HTNV) and Seoul virus (SEOV) are two major causative agents of HFRS in China (Jonsson et al., 2010; Zhang et al., 2010). Clinical manifestations are vast, ranging from acute febrile, mucocutaneous petechia illness to the gravis form, which occurs when patients develop hemorrhage and shock due to increased vascular permeability (Liu et al., 2009; Jonsson et al., 2010; Zhang et al., 2010).
The pathogenesis of HFRS is complex and involves capillary leakage due to infection of vascular endothelial cells (Gavrilovskaya et al., 1999; Mackow and Gavrilovskaya, 2001). Virus receptors are cell surface molecules that bind incoming viruses to the cells, promote their entry, and even trigger signaling cascades in the host cells. Integrins, composed of noncovalently associated α (n=18) and β (n=8) subunits, comprise a large family of heterodimeric transmembrane glycoproteins and can mediate cell adhesion and binding to the extracellular matrix (Hynes, 2002). β3 integrins, including αVβ3 and αIIbβ3 integrins, are expressed on endothelial cells, macrophages, platelets, and certain activated leukocytes. Previous studies have demonstrated that integrins can mediate the cellular entry of pathogenic hantaviruses into endothelial cells, resulting in pathological consequences, such as alterations to vascular permeability, cell integrity, and hemostasis. Endothelial dysfunction caused by pathogenic hantaviruses can have various effects on vascular permeability via endothelial and immune cell responses, leading to multifactorial disease pathogenesis (Mackow and Gavrilovskaya, 2001; Gavrilovskaya et al., 2002).
Single nucleotide polymorphisms (SNPs) are genetic markers that are arguably the most important type of variation in the genome and are responsible for genetic effects that produce susceptibility to many human diseases. A number of promising SNP genotyping methods are currently available and have been applied to explore the genetic determinants of common disease (Syvanen, 2001; Kwok and Chen, 2003). Previous studies have reported that SNPs in integrins β3, αV, and αIIb are associated with human diseases such as asthma, atherosclerosis, and rheumatoid arthritis (RA) (Jacq et al., 2007; Kucharska-Newton et al., 2011; Zhang et al., 2013). Liu et al. demonstrated that HPA-3 polymorphisms (integrin αIIbβ3) are significantly different in patients with HFRS, and the frequency of the HPA-3 b allele increases with disease severity (Liu et al., 2009). However, the relationship between SNPs in integrin αvβ3 and HFRS disease progression has not been reported.
Thus, in this study, we investigated genetic polymorphisms in five individual sites of the integrin αvβ3 gene in 90 patients with HFRS and 101 normal individuals, using TaqMan SNP Genotyping Assays and by bidirectional PCR allele-specific amplification (Bi-PASA) test. The associations between gene polymorphisms in αvβ3 and HFRS disease susceptibility and severity were analyzed, which would further illuminate the potential role of integrin αvβ3 in the pathogenesis of HFRS in Hubei Province.
-
Ninety patients (51 men and 39 women; mean age: 42±10.0 years) diagnosed with HFRS were enrolled in this study. Patients were recruited from 12 districts in Hubei Province, China and were unrelated. All patients were diagnosed during hospitalization from 1986 to 1995 according to standard clinical criteria (Xiong et al., 2011), which mainly include: age≥14 years, febrile time≤4 days, typical HFRS symptoms and signs (e.g., fever, proteinuria, hematuria), and potential exposure history to wild rodents or other HFRS patients. Patient sera were subsequently confirmed to be positive by indirect IgM/IgG enzyme-linked immunosorbent assays (ELISAs) performed in our laboratory, using mixed antigens of HTNV and SEOV. Other kidney disorders, diabetes, cardiovascular and hematologic disease, viral hepatitis, and autoimmune disease were excluded.
The control subjects included 101 healthy, unrelated blood donors (56 men and 45 women; mean age: 39±11.0 years) without a history of HFRS-like disease recruited from hospital. Both patients and controls were of Han Chinese descent. There were no significant differences in the distributions of ages and sexes between patients with HFRS and healthy controls. Written informed consent was obtained from all patients and controls in the study. The Research Ethics Committee of Wuhan University approved this project.
Patients with HFRS were classified into four clinical types (mild, medium, severe, and gravis), according to the diagnostic criteria outlined in Prevention and Treatment Strategy of HFRS (Ministry of Health, People's Republic of China, 1997; Liu et al., 2008b). Criteria for classification of types included severity of toxemic symptoms, blood pressure, extent of hemorrhage, severity of kidney damage (proteinuria occurrence and duration of oliguric period as well as levels of urea nitrogen), and the occurrence and severity of complications. Only 70 patients were included in this analysis, as clinical data for disease classification were not available for the remaining 20 patients.
-
Blood samples were collected in ethylene diamine tetra acetic acid (EDTA)-anticoagulant tubes and stored at -80 ℃ until use. Total genomic DNA was extracted from 500-1000 μL of blood by standard proteinase K/phenol methods (Ahmad et al., 1995). All DNA samples were quantified using a NANODROP 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and stored at -20 ℃ until use.
-
SNP genotype data for Han Chinese in Beijing, China (CHB) were obtained from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/snp) and HAPMAP database (http://snp.cshl.org/cgi-perl/gbrowse/hapmap27_B36/). A total of five SNPs were selected for genotyping: two for the intron regions of ITGAV (which encodes an α chain integrin subunit, located at chromosome 2) and three for the exon regions of ITGB3 (which encodes a β chain integrin subunit, located at chromosome 17). The two intronic SNPs from the 5′ and 3′ ends of ITGAV were rs3768777 (A/G) and rs3738919 (A/C); and the three exonic SNPs with high allele frequencies from ITGB3 were rs5918 (C/T, exon 10), rs13306487 (A/G, exon 10), and rs5921 (A/G, exon 3). Detailed information of the selected SNPs in this study is given in Table 1.
Gene ITGAV ITGB3 SNP rs3768777 rs3738919 rs5918 rs5921 rs13306487 Location in gene Intron Intron Exon Exon Exon Position 37665539 37730678 4014008 4022879 4023066 Location in mRNA - - 196 1377 1564 Nucleotide base A-G A-G C-T A-G A-G Amino acid mutation - - Pro-Leu Ile-Val Gln-Arg MAF in CHB 0.232 0.058 0.007 0.004 0.050 Table 1. Background information of selected SNPs in this study
-
TaqMan SNP genotyping assays and Bi-PASA test were employed to amplify the target regions. Genotyping of rs5918 was conducted on DNA specimens according to the TaqMan Genotyping Assay protocol, using 5′ nuclease assays with allele-specific TaqMan probes (Applied Biosystems, Foster City, CA, USA) on a Bio-Rad CFX96 instrument (Bio-Rad, Hercules, CA, USA). The reaction protocol was as follows: 95 ℃ for 10 min; 40 cycles of 92 ℃ for 15 s and 60 ℃ for 1 min; and 72 ℃ for 5 min. Genotyping of rs3768777, rs3738919, rs13306487, and rs5921 was conducted by Bi-PASA test, since the SNP sites were located in single nucleotide mutation-rich regions, which are not easily analyzed by TaqMan SNP genotyping assays. Bi-PASA test was also performed because it involves fewer steps, costs less, and is easier to interpret (Liu et al., 1997). The amplification was carried out according to the following protocol: initial denaturation at 95 ℃ for 3 min; 95 ℃ for 30 s, 30 s touchdown from 68 ℃ to 60 ℃ in 0.8 ℃ steps, followed by 26 cycles at 60 ℃ for 30 s and at 72 ℃ for 30 s; and a final extension at 72 ℃ for 5 min. PCR primers used for rs3768777, rs3738919, rs13306487, and rs5921 SNPs amplification are listed in Table 2.
SNP Primers rs3768777 rs3768777-p 5′-GTTGCTAATGTTCCGCGTTGCA-3′ rs3768777-q 5′-GTAGTAGAAGATGGTCCTATCCACG-3′ rs3768777-a 5′-GGGGGGGGGCTCATCACCCCACCCCCA-3′ rs3768777-b 5′-GGGGGGGGCGTGCTCCTAACGCTAACAT-3′ rs3738919 rs3738919-p 5′-ATTTCCAGGTGGAACTTCTTTTGGA-3′ rs3738919-q 5′-TCACAATTCAGATTTTTGCCACTGG-3′ rs3738919-a 5′-GGGGGGGGGCGACACAAAGGAAATTTAGA-3′ rs3738919-b 5′-GGGGGGGGGCGGTGTGACACTTTACAAAG-3′ rs13306487 rs13306487-p 5′-AAGGCTGAGGAACTCCAGATTG-3′ rs13306487-q 5′-TGTTTCCAGTGGTTGCAGGTAT-3′ rs13306487-a 5′-GGGGGGGGGCCGAATGCAGCCCCCA-3′ rs13306487-b 5′-GGGGGGGGGCACGGGCTGACCCTCCC-3′ rs5921 rs5921-p 5′-AAGGCTGAGGAACTCCAGATTG-3′ rs5921-q 5′-TGTTTCCAGTGGTTGCAGGTAT-3′ rs5921-a 5′-GGGGGGGGGCTCAAGGACAGCCTGATCA-3′ rs5921-b 5′-GGGGGGGGGCATCAAAGGTGACCTGGAC-3′ Table 2. PCR primers used for SNPs amplification
The amplified fragments were readily distinguishable by electrophoresis through a 2% agarose gel. The results obtained for the genotypes were further validated by sequencing analysis of 10 randomly selected samples (data not shown).
-
Statistical analysis was performed using SPSS 17.0 software. The genotypic frequencies of all SNPs in the different groups were calculated, and their distributions were analyzed by Hardy-Weinberg equilibrium statistics. Genotypic and allelic frequencies between the HFRS and control groups were assessed by χ2 analysis. Genotype and allele distributions between clinical types were analyzed by Mann-Whitney U tests. Differences with P values of less than 0.05 were considered statistically significant.
Study participants
Blood collection and DNA isolation
SNP selection and primer design
Genotyping of candidate SNPs of integrin αvβ3
Statistics analysis
-
All the genotype frequencies of the studied SNPs were subjected to Hardy-Weinberg Equilibrium (HWE) statistics. The observed values of candidate SNP genotype frequencies in each group demonstrated no significant differences compared with the corresponding expected values in each group (P > 0.05). Genotype distributions in each group did not deviate from HWE and were in accordance with genetic equilibrium.
-
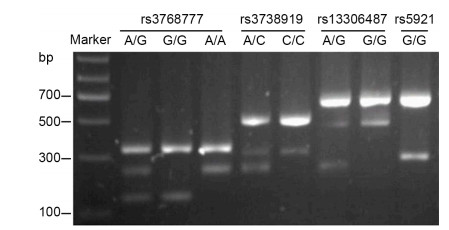
The rs5918 SNP was detected by TaqMan SNP genotyping assays, as described above, and the C/T or T/T genotype was represented in different amplification curves by real-time quantitative PCR (data not shown). As shown in Figure 1, Bi-PASA test produced two fragments for the homozygote and three fragments for heterozygote.
-
The distributions of genotypes and allelic frequencies of four candidate SNPs of integrin αvβ3 (rs5918, rs3768777, rs3738919, and rs13306487) were comparable between patients with HFRS and controls (all P > 0.05; Table 3). In addition, there was only one genotype of the rs5921 locus (G/G) detected both in healthy individuals and patients with HFRS (Figure 1 and Table 3). There were three genotype variants of rs3768777 (A/G, G/G, and A/A) in the HFRS and control populations; however, the allelic frequencies between groups were equivalent (P > 0.05). For rs5918, rs3738919, and rs13306487, two types of mutant alleles were observed. The majority were homozygous (T/T, C/C, and G/G genotypes, respectively) with similar distributions in patient and control cohorts (all P > 0.05).
SNP Group Genotypes Allele frequency (%) aa ab bb χ2 P a b χ2 P rs5918 Controls 0 1 100 0.467 0.494 1(0.5) 201(99.5) 0.464 0.496 Patients 0 2 88 2(1.1) 178(98.9) rs3768777 Controls 6 35 60 0.354 0.838 47(23.3) 155(76.7) 0.000 0.988 Patients 4 34 52 42(23.3) 138(76.7) rs3738919 Controls 0 14 87 0.328 0.567 14(6.9) 188(93.1) 0.306 0.580 Patients 0 10 80 10(5.6) 170(94.4) rs13306487 Controls 0 6 95 0.721 0.396 6(3.0) 196(97.0) 0.073 0.402 Patients 0 3 87 3(1.7) 177(98.3) rs5921 Controls 0 0 101 - - - - - - Patients 0 0 90 Table 3. Comparison of genotypes and allele frequencies of candidate SNPs between patients with HFRS and controls
-
We further analyzed the associations between two more highly detectable SNP sites and disease severity of HFRS. Table 4 illustrated the demographic and clinical characteristics of 70 of the patients included in this analysis. The numbers of patients with mild, moderate, severe, and gravis types were 9, 49, 11, and 1, respectively. As shown in Table 5, the distributions of genotypes at rs3768777 and rs3738919 sites among patients with difference clinical types of HFRS did not differ significantly (all P > 0.05), nor did the allele frequencies of a and b.
Clinical features and laboratory data Mild type (n=9) Medium type (n=49) Severe type (n=11) Gravis type (n=1) Age (years)a 25.6±6 34.2±12.9 36.6±18.3 17 Sex (man/women) 7/2 38/11 7/4 1/0 WBC count at admission (×109/L) < 10 4 14 2 ≥10 5 35 9 1 Platelet count at admission (×109/L) ≥50 4 12 1 < 50 5 37 10 1 Maximum serum creatinine at oliguric phase (μmol/L)a 221.6±83.1 434.6±243.0 698.6±308.6 760.4 Patient hospital duration (days)a 10.7±2.2 12.0±4.6 15.6±12.3 2.0 Note: aData are presented as means±SDs Table 4. Demographic, clinical, and hematochemical characteristics in patients with different clinical types of HFRS
SNP Clinical type Genotypes Allele frequency (%) aa ab bb a b rs3768777 Mild 1 1 7 3(8.6) 15(14.3) Medium 3 19 27 25(71.5) 73(69.5) Severe 0 6 5 6(17.1) 16(15.2) Gravis 0 1 0 1(2.8) 1(1.0) rs3738919 Mild 0 2 7 2(25.0) 16(12.1) Medium 0 4 45 4(50.0) 94(71.2) Severe 0 2 9 2(25.0) 20(15.2) Gravis 0 0 1 0(0.0) 2(1.5) Table 5. Genotypes and allelic frequencies of rs3768777 and rs3738919 in the integrin αvβ3 gene in patients with different clinical types of HFRS
Genetic equilibrium analysis of candidate SNPs of integrin αvβ3
Detection of amplicons
Frequency distributions of candidate SNPs of integrin αvβ3 alleles between patients with HFRS and normal controls
Distributions of candidate SNPs in the integrin α vβ3 gene and allele frequencies in patients with HFRS with different clinical types
-
Integrin αvβ3 has been proposed to be the cellular receptor for pathogenic hantaviruses and thus may play a pivotal role in the pathogenesis of HFRS (Gavrilovskaya et al., 1999; Mackow and Gavrilovskaya, 2001). Previous studies have confirmed the association of polymorphisms in integrin β3, αv, and αIIb with certain diseases, including asthma, atherosclerosis, giant thrombocytopenia, and rheumatoid arthritis (Jacq et al., 2007; Kucharska-Newton et al., 2011; Zhang et al., 2013). However, no reports are available concerning the relationship between SNPs of α vβ3 and HFRS in Han Chinese individuals.
In the present study, TaqMan SNP genotyping assays and Bi-PASA test were applied to genotype the selected SNPs in ITGB3 and ITGAV genes. Five SNPs were accurately and successfully genotyped. Based on the genotyping results, the association between the five SNPs in the integrin αvβ3 gene and HFRS were investigated in Han Chinese individuals from Hubei Province.
Three ITGB3 SNPs (rs5918, rs13306487, and rs5921) were selected for this study because these SNPs have the highest allele frequencies in the Han population database. However, our results suggested that these SNPs were not associated with HFRS in the Chinese Han population. Similar results have been observed in previous studies of these SNPs in other diseases. The rs5918 variation was shown to be associated with wheezing and asthma in the Madison population (Thompson et al., 2007), but no relationship was observed in Chinese Han children (Zhang et al., 2013). Jacq et al. (2007) concluded that the rs3738919-C allele of the ITGAV gene, rather than the rs3768777 SNP, is associated with RA in the European Caucasian population, with the C allele increasing the risk of illness. Our results, however, did not suggest that these two ITGAV SNPs were HFRS susceptibility gene sites and did not support a role for these two SNPs in HFRS disease severity.
Liu et al. (2008a) found that the intensity levels of platelet membrane β3 integrin were associated with clinical phases and disease diversity in patients with HFRS in the neighbor province of Shaanxi, China. They further showed that SNPs in integrin αIIbβ3 were associated with susceptibility to hantavirus infection and the disease severity of HFRS (Liu et al., 2009). With regard to Puumala virus infection, Laine et al. (2012) found that HPA-1(integrin αIIbβ3, rs5918) did not correlate with disease severity in Finland. These differences may be related to regional diversity, differences in viral strains, ethnic variations, and other factors. Our results did not support the concept of a potential relationship between SNPs in integrin αvβ3 and HFRS disease progression. Thus, we presume that we were unable to confirm the associations of these five SNPs in the α vβ3 gene with susceptibility to or severity of HFRS in Hubei Province because of differences between the Han population and other ethnicities, low sample size, and environmental effects. Nonetheless, it should be noted that we did not examine SNPs covering a maximum amount of genetic variation in the ITGB3 and ITGAV genes, and thus, we could not exclude the possibility that SNPs located in other regions of the ITGB3and ITGAV genes may be involved in the development of HFRS in Hubei Province.
In summary, our results did not provide support for associations of rs5918, rs13306487, rs5921, rs3738919, and rs3768777 with HFRS, either as single susceptibility gene sites or determination factors for disease severity. Although the presence of other risk variants in ITGB3 and ITGAV genes cannot be excluded, the present study is the first step in exploring the relationship between integrin αvβ3 gene polymorphisms and HFRS among the Han population in Hubei Province. However, further studies with more patients are needed to draw firmer conclusions as to whether α vβ3 gene polymor-phisms are related to the risk of HFRS.
-
We thank the physicians who assisted with sample collection and Dr. Jing Sun, School of Public Health, Wuhan University, for statistical assistance. We thank Dr. RheaBeth Markowitz, Georgia Regents University, USA, for editing assistance. This work was supported by grants from the National Natural Science Foundation of China (grant nos. 81101258, 81000734, and 81271819) and a grant from Hubei Province Health and Family Planning Scientific Research Project (grant no. WJ2015MB113).
-
All the authors declare that they have no conflicts of interest. Informed consent was obtained from all patients for whom identifying information is included in this article.
-
XPC, HRX, and WH conceived and designed the research. XPC, HRX, NZ, and QZC carried out the experiments. XPC, HW, CJZ, MRW, SL, and FL analyzed the data. XPC, HRX, and WH wrote the paper. All authors have read and approved the final manuscript.














 DownLoad:
DownLoad: