-
Dear Editor,
Severe fever with thrombocytopenia syndrome virus (SFTSV) is a newly identified viral pathogen of the genus Phlebovirus in the family Bunyaviridae (Sun et al., 2012). SFTSV was first identified from patient serum samples in China (Li et al., 2013; Ning et al., 2015). SFTSV can cause a severe hemorrhagic fever-like disease with a reported case fatality rate ranging from 2.5% to 30%, representing a significant public health problem, (Liu et al., 2013). However, the pathogenesis of STFSV remains unknown, and there are no vaccines or specific antivirals available (Ning et al., 2015). The SFTSV genome consists of three negative single-stranded RNA segments, including the L, M, and S segments (Yu et al., 2011). The S segment encodes the nucleoprotein (NP), which was shown to be highly immunogenic and is conserved among all isolates of phleboviruses (Xu et al., 2011; Martin et al., 2010; Fukushi et al., 2012). Antibodies against the SFTSV NP are commonly detected early after infection, suggesting that the NP is a candidate target antigen for the early diagnosis of SFTSV infection (Li, 2011). Furthermore, Liu et al. (2011) reported a high infection rate among domestic animals, and Ge et al. (2012) observed SFTSV infections in rodents. However, the host ranges of SFTSV are still not clear. According to previous studies, there is an epidemic of CrimeanCongo hemorrhagic virus, which is a bunyavirus, in the Xinjiang region, although it is not clear whether SFTSV infection exists in potential host vertebrates (Wei et al., 2010; Sun et al., 2011). Therefore, establishing methods for the accurate and rapid diagnosis of SFTSV in host animals is an urgent requirement. Recombinant antigens have thus far been used for the accurate and specific detection of antibodies of a number of viruses in the family Bunyaviridae (Jansen et al., 2007). In the present study, we used the recombinant NP of SFTSV to establish an indirect ELISA method for the specific, sensitive, safe, and rapid detection of SFTSV.
The NP gene of SFTSV (HB29 strain) was amplified and inserted into the expression plasmid pET-28a by restriction endonuclease digestion with Hind Ⅲ and EcoR Ⅰ. After being transformed into competent Escherichia coli BL21 cells, the recombinant plasmid pET-28a-SFTSVNP was identified by restriction enzyme digestion (Supplementary Figure S1A). DNA sequence analysis (Sangon Biotech, Shanghai, China) showed that the fulllength sequence of the constructed NP gene was 744 bp, which was completely consistent with that of the SFTSV HB29 strain (NC_018137.1). These results suggested that the recombinant plasmid pET-28a-SFTSV-NP was correctly constructed.
Expression of the recombinant plasmid pET-28aSFTSV-NP was induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG). SDS-PAGE analysis of the expressed product SFTSV-rNP revealed a specific band at approximately 28 kDa after induction, which was not detected in the non-induced culture, and the SFTSV-rNP was mostly expressed in inclusion bodies rather than in the culture medium (Figure 1A-a). These results indicated that SFTSV-rNP was successfully expressed.
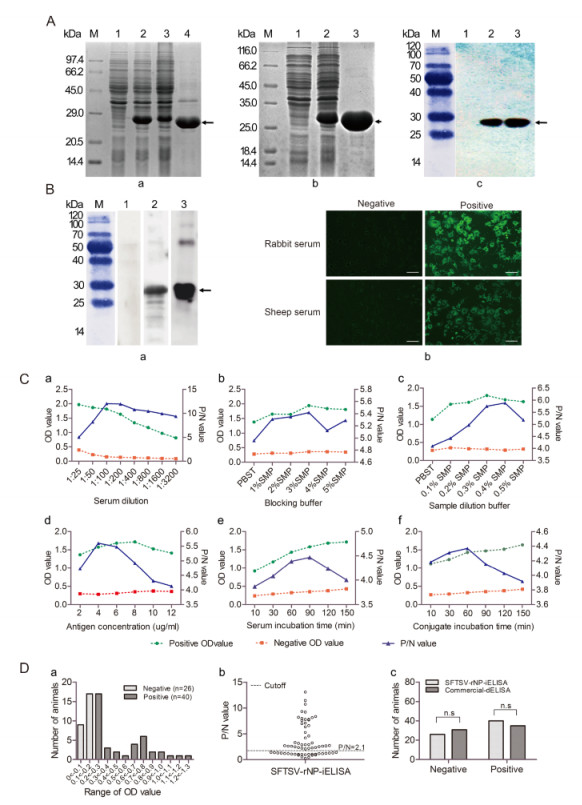
Figure 1. (A) Expression, purification, and identification of SFTSV-rNP. (A-a) SDS-PAGE analysis of SFTSV-rNP expression. M: standard protein marker; lanes 1 and 2, lysates before and after induction; lanes 3 and 4, supernatant and inclusion bodies. (A-b) The purification of SFTSV-rNP. Lanes 1 and 2, before and after induction; lane 3, purified SFTSV-rNP. (A-c) Identification of SFTSV-rNP by Western blot. Lanes 1 and 2, lysates before and after induction; lane 3, purified SFTSV-rNP. (B) The antigenicity and reactivity of SFTSV-rNP and sera. (B-a) Identification of the antigenicity of SFTSV-rNP by Western blot. Lane 1, negative sheep serum; lane 2, positive sheep serum; lane 3, positive control (rabbit serum). (B-b) Identification of the reactivity of sera with the viral protein by IFA; bars indicate 50 μm. (C) Optimization of SFTSV-rNP-iELISA conditions. The suitable conditions were determined with a serum sample dilution of 1:100 (C-a), 3% SMP as the blocking buffer (C-b), 0.4% SMP as the sample dilution buffers (C-c), 4 μg/mL as the SFTSV-rNP package concentration (C-d), 30 min of serum incubation time (C-e), and 60 min of conjugate incubation time (C-f). (D) Detection of serum samples by ELISA. (D-a) Distribution of OD values for each sheep serum sample by SFTSV-rNPiELISA; negative sera (n=26) and positive sera (n=40). (D-b) Distribution of P/N values for each serum sample; the cutoff value was P/N=2.1. (D-c) Comparison of SFTSV-rNP-iELISA and commercial dELISA results; there was no statistically significant difference between the methods in the numbers of positive and negative serum samples (χ2=0.773, P=0.379).
The conditions of protein expression and purification were optimized to obtain the recombinant protein with high quantity and purity. To improve the protein productivity, the induction temperature, concentration of inducer IPTG, initial bacteria concentration and induction time were determined. Expressed proteins in different conditions were analyzed by SDS-PAGE. Comparison of samples exposed to five different temperatures, from 28 ℃ to 45 ℃, showed that the highest expression level of the fusion protein was obtained at 37 ℃ (Supplementary Figure S1B-a). The optimized concentration of the inducer IPTG was 0.2 mmol/L (Supplementary Figure S1B-b), and the initial bacteria concentration was determined with an optical density at 595 nm (OD595) value from 0.4 to 0.8 (Supplementary Figure S1B-c). The highest yield of the expressed SFTSV-rNP was detected at 4 h after induction (Supplementary Figure S1B-d). To optimize the purification condition using Ni affinity chromatography, the expressed SFTSV-rNP from the inclusion bodies were added to a Ni affinity chromatography column and reacted for 3 h at 4 ℃. After the column was washed sequentially with increasing concen-trations of imidazole from 20 mmol/L to 500 mmol/L for optimization, the concentration of 120 mmol/L imidazole was determined to be optimal for washing the target protein (Supplementary Figure S1C). The expressed SFTSV-rNP in inclusion bodies was purified as described above. The SDS-PAGE results revealed a single pure band with the expected size (Figure 1A-b). Gray-scale scanning analysis showed that the purification rate of the fusion protein reached up to 91.26%, suggesting that the expression and purification procedures of SFTSV-rNP established in this study were efficient.
The identity of the expressed and purified SFTSV-rNP was further confirmed by Western blot analysis. A mouse anti-His antibody (1:2000) was used as the primary antibody, and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (TransGen Biotech, Beijing, China) was used as the secondary antibody. The results showed that the mouse anti-His antibody was recognized specifically by the induced culture and purified SFTSV-rNP, but not by the non-induced culture (Figure 1A-c), suggesting that SFTSV-rNP was expressed correctly.
To confirm the antigenicity of SFTSV-rNP and the accuracy of the serum test results, Western blot analysis and an immunofluorescence assay (IFA) were carried out. SFTSV-rNP was first separated by SDS-PAGE and then transferred to nitrocellulose membranes; SFTSVnegative or -positive sheep or rabbit (control) sera (1:100) were used as the primary antibody, and HRP-conjugated goat anti-rabbit or rabbit anti-sheep IgG (1:4000; TransGen Biotech) was used as the secondary antibody. The target bands were visualized and imaged by GE-ImageQuant-LAS-4000 (Japan). The western blot results showed that SFTSV-rNP had an apparent effect on binding to SFTSV-positive sheep and rabbit sera, but not SFTSV-negative sera (Figure 1B-a). The IFA was conducted to further confirm the reactivity of the antibody in the sheep serum with the viral particle. For this analysis, Vero-E6 cells infected with SFTSV (HB29 strain) were used, with sheep or rabbit sera (1:100) as the primary antibody and fluorescein isothiocyanate-labeled goat antirabbit IgG or rabbit anti-goat IgG (1:2000) as the secondary antibody, finally the effects were visualized under an immunofluorescent microscope (TI-SH-J, Nikon, Japan). The IFA results showed that the SFTSV-positive rabbit and sheep sera were clearly reactive with the SFTSV viral protein, but the negative sera were not (Figure 1B-b). These Western blot and IFA results suggest that SFTSVrNP has efficient antigenicity.
The optimal conditions of SFTSV-rNP-based indirect ELISA (SFTSV-rNP-iELISA) were established, including the serum sample dilution, blocking buffer, sample dilution buffer, antigen concentration, serum sample exposure time, and conjugating time. At first, the purified SFTSV-rNP was coated at 200 μL/well on 96-well microtiter plates with a concentration ranging from 2 to 10 μg/mL. On the basis of the results described above, SFTSV-positive and -negative control sera were also diluted in serial dilutions from 1:25 to 1:3200 for optimization. After application, the optimal serum sample dilution and antigen concentration were set at 1:100 and 4 μg/mL, respectively (Figure 1C-a, 1C-d), since these conditions yielded the highest OD450/630 ratio between positive and negative sera. Thereafter, the blocking and sample dilution buffers were optimized, and a 3% skimmed milk powder (SMP) in phosphate buffered saline with Tween (PBST) was established as the blocking buffer and 0.4% SMP was established as the optimized sample dilution buffer (Figure 1C-b, 1C-c). After the above conditions were determined, the optimal exposure times of the serum samples and conjugate were optimized such that incubations of 30 min and 60 min were carried out for serum samples and the conjugate, respectively, at 37 ℃ (Figure 1C-e, 1C-f). All these conditions were sufficient and time-saving for carrying out SFTSVrNP-iELISA, and the measured OD450/630 values were relatively stable and were confirmed to improve the accuracy of the verification.
A total of 66 sheep serum samples (CDC, Xinjiang) were tested using the established SFTSV-rNP-iELISA with the optimized conditions. The purified SFTSV-rNP was diluted to 4 μg/mL. The 96-well microtiter ELISA plates were coated with SFTSV-rNP (200 μL/well) and then blocked with 300 μL/well of 3% SMP in PBST, followed by incubation with 100 μL/well of sheep serum (1:100), and subsequently incubated with HRP-conjugated rabbit anti-sheep IgG (1:2000). The absorbance was measured at an incidence wavelength of 450 nm and a reference wavelength of 630 nm in the microplate reader (Bio Tek, USA). The test results are presented as the frequency distribution of the OD450/630 ratio of each serum sample. The OD450/630 ratios of the negative sera were in the range of 0-0.2, whereas those of the positive sera were in the range of 0.2-1.3 (Figure 1D-a). In this study, SFTSV-rNP-iELISA was performed with a P/N ratio (P/N=mean of each tested sample OD450/630/mean of the OD450/630 of the negative control) as the cutoff value to distinguish between the positive and negative results. If the P/N > 2.1, the results were considered to be positive, and were otherwise considered to be negative (Figure 1D-b). The results showed that 26 of the 66 serum samples were classified as negative and 40 of the 66 serum samples were classified as positive in SFTSV-rNP-iELISA.
To determine the sensitivity and specificity of the SFTSV-rNP-iELISA, all of the sheep serum samples were further tested using a commercial double-antigen sandwich ELISA kit, and the results of a portion of the sheep serum samples were additionally confirmed by IFA and Western blot (data not shown). Overall, 35 of the 66 samples were detected to be SFTSV-rNP-positive, and 31 samples were detected to be negative with the commercial dELISA kit, and 35 positive and 26 negative samples were detected by both methods (Table 1). In the detection, the specificity, sensitivity, and concordance of SFTSV-rNP-iELISA were 100%, 87.5%, and 92.42%, respectively (Table 1). There was no significant difference (P=0.379) between the numbers of positive and negative serum samples detected with the SFTSVrNP-iELISA and commercial dELISA kit (Figure 1D-c). These results indicated that the established indirect rNPELISA method has efficient sensitivity and specificity.
SFTSV-rNP-iELISA Commercial dELISA kit Total positive negative Positive 35 5 40 Negative 0 26 26 Total 35 31 66 Note: Specificitya: 100%; Sensitivityb: 87.5%; Concordancec: 92.42% aTrue negative/(False positive + True negative)=26/(0 + 26)=100% bTrue positive/(False negative + True positive)=35/(35 + 5)=87.5% c(Positive by both methods + Negative by both methods)/Total number of samples=(35 + 26)/66=92.42%. Table 1. Comparison of SFTSV-rNP-iELISA and commercial double-antigen sandwich ELISA kit for antibody detection
In conclusion, SFTSV-rNP was successfully expressed and purified, and its antigenicity was identified by Western blot. A total of 66 sheep serum samples were investigated using the newly developed SFTSV-rNP-iELISA method. The coincidence ratio between the commercial double-antigen sandwich ELISA kit for detecting SFTSV IgG antibody and the established indirect ELISA method reached up to 90.45%. The developed indirect ELISA method which had higher specificity and sensitivity, is safe and easy to operate, and is therefore suitable for epidemiological investigations and the diagnosis of SFTSV. At present, there are no reported SFTSV infection cases in humans in the Xinjiang region. Our preliminary results showed that the serum of a domestic animal (sheep) shows a high positive rate for SFTSV antibody, indicating that there may be a risk of SFTSV infection in this region, highlighting the importance of monitoring SFTSV infections in domestic animals. More theoretical research and knowledge is needed to prevent and control the infection and outbreak of SFTSV. The results of the present study lay a foundation for further research and development of SFTSV epidemiological investigations and establishment of an ELISA diagnostic kit.
HTML
-
This work was supported by the grants from the National Science Foundation of China (No. 81460303), the Ministry of Science and Technology of China (2013FY113500), and Funded by the Open Research Fund Program of the State Key Laboratory of Virology of China (No. 2015IOV003). The authors declare that they have no conflict of interest.
Supplementary Figure S1 is available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
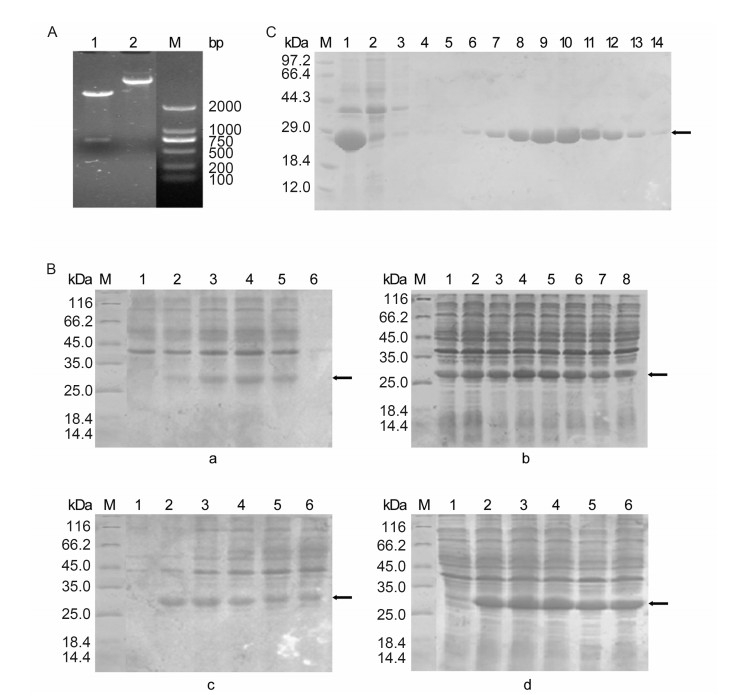
Figure S1. (A) Analysis of the recombinant plasmid pET-28a-SFTSV-NP by digestion. Lane 1, the plasmid pET-28aSFTSV-NP; Lane 2, the plasmid pET-28a-SFTSV-NP digested by Hind Ⅲ and EcoR I; M: DNA marker. (B) SDS-PAGE analysis of SFTSV-rNP expression in different conditions. (B-a) Effect of induction temperature on protein expression. M: standard protein marker; lane 1: before induction; lanes 2, 3, 4, 5, and 6: induction temperature of 28 ℃, 32 ℃, 37 ℃, 42 ℃, and 45 ℃, respectively. (B-b) Effect of IPTG concentration on protein expression. Lanes 1, 2, 3, 4, 5, 6, 7, and 8: IPTG concentration of 0.02 mmol/L, 0.05 mmol/L, 0.1 mmol/L, 0.2 mmol/L, 0.4 mmol/L, 0.6 mmol/L, 1 mmol/L, and 2 mmol/L, respectively. (B-c) Effect of initial bacterial concentration on protein expression. Lane 1: before induction; lanes 2, 3, 4, 5, and 6: OD value of bacterial concentration of 0.286, 0.551, 0.851, 1.038, and 1.210, respectively. (B-d) Effect of induction time on protein expression. Lane 1: before induction; lanes 2, 3, 4, 5, and 6: induction time of 2 h, 3 h, 4 h, 5 h, and 6 h, respectively. (C) SDS-PAGE analysis of recombinant SFTSV-NP purification in different washing concentrations. M, protein marker; lane 1, total protein extraction; lane 2, uncombined protein; lanes 3-14, eluted fraction from column which was washed sequentially with increasing concentrations of imidazole (20 mmol/L, 20 mmol/L, 20 mmol/L, 40 mmol/L, 60 mmol/L, 80 mmol/L, 100 mmol/L, 120 mmol/L, 140 mmol/L, 160 mmol/L, 180 mmol/L, and 200 mmol/L).
















 DownLoad:
DownLoad: