HTML
-
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging disease caused by a novel tick-borne virus, named severe fever with thrombocytopenia syndrome bunyavirus (SFTSV) (Yu et al., 2011). SFTS patients usually develop symptoms including severe fever, thrombocytopenia, leucopenia, central nervous system symptoms, and gastrointestinal symptoms, which lead to multiple organ failure or even death (Li, 2013). SFTSV belongs to the family Bunyaviridae, genus Phlebovirus. SFTSV is an enveloped virus enclosed with negative single-stranded RNA composed of three RNA segments, the large (L) segment encoding the RNA-dependent RNA polymerase (RdRP), the medium (M) segment encoding a precursor of glycoprotein (G), and the small (S) segment encoding a nucleoprotein (NP) and a nonstructural protein (NSs) using an ambisense coding strategy (Yu et al., 2011; Jiao et al., 2012).
SFTS disease has been reported in at least 13 provinces in China. Epidemiological investigations and evaluations of environmental risk factors have found that hilly areas, including forests, shrubs, and rain-fed croplands, in provinces located in Central and Eastern China are strongly associated with a high risk of SFTS outbreaks (Xu et al., 2014; Wu et al., 2016). These SFTSV outbreaks were first reported in 2009 and 2010 in Huaiyangshan Mountain area, which lies at the border of Hubei, Henan, and Anhui Provinces and covers the cities close to Tongbaishan Mountain and Dabieshan Mountain, including Xinyang in Henan Province, and Suizhou, Xiaogan, and Huanggang in Hubei Province (Yu et al., 2011; Zhang et al., 2011). Therefore, these areas have been targets for disease prevention and control. Surveys conducted by a hospital in Xinyang City, Henan Province, have confirmed hundreds of SFTS cases in recent years (Xu et al., 2014), and a number of SFTSV strains have been isolated from patients. However, relatively fewer studies have been performed in SFTS patients in the neighboring areas of Hubei Province. Epidemiological investigations conducted in Hubei Province have identified SFTSV RNA in serum samples of villagers, animals, and ticks by reverse transcription polymerase chain reaction (RT-PCR) and also found positive antibody in serum samples from villagers by serological analyses (Xing et al., 2016). Additionally, cases of SFTS from Hubei have been described in a few reports (Yu et al., 2011; Zhang et al., 2011; Chen et al., 2013).
To date, four SFTSV strains (HB29, HB154, HB155, and HB156) have been isolated from serum samples of patients with SFTS, and the complete genome sequences were deposited in GenBank for further studies (Yu et al., 2011). Phylogenetic analysis with the complete genome sequences of SFTSV strains deposited in GenBank showed that SFTSV could be classified into five genotypes in association with geographic distribution, including C1 to C4, containing almost all strains from China, and J, containing strains from Korea and Japan (Yoshikawa et al., 2015; Shi et al., 2016). Moreover, the strains from Hubei Province were found to belong to one clade (the C3 clade), suggesting that a simple genotype of SFTSV has been circulating in Hubei Province (Yoshikawa et al., 2015; Shi et al., 2016). However, these results were based on very limited Hubei isolates and might not reflect the real situation.
In this study, we report three SFTS cases identified in 2015 and 2016 from two different regions in Hubei Province. Three new SFTSV strains were isolated from the serum samples. Furthermore, the virus properties were characterized, and the genomes were completely sequenced. Phylogenetic analyses found that the three strains belonged to two different genotypes, which revealed that SFTSVs of more than one genotype have been circulating in Hubei Province.
-
Three patients who visited the Department of Infectious Disease in Union Hospital had typical SFTS symptoms. The patients underwent routine blood tests, urine routine tests, and serum biochemistry tests. Serum samples were collected from the three patients on the first day of admission. An SFTSV quantitative RT-PCR (qRT-PCR) detection kit (Daan Gene, China) was used to detect SFTSV RNA in serum samples from hospitals according to the manufacturer's instructions.
SFTSV RNA-positive serum samples were used for virus isolation by blind passages. Briefly, at 40% confluence, Vero cells were seeded in six-well plates, incubated for 1 day, and then incubated with 50 μL serum diluted in Dulbecco's modified Eagle's medium (DMEM; Sigma, USA) with 2% fetal bovine serum (FBS; Gibco, Australia) at 37 ℃ for 1 h. The supernatants were then replaced with fresh DMEM with 2% FBS containing penicillin (50-100 IU/mL) and streptomycin (50-100 μg/mL) and incubated at 37 ℃ for 7 days. Supernatants from each sample were harvested and used to incubate with new healthy cells. After three passages, the supernatants from each sample were harvested to remove cell debris by centrifugation (3000 × g, 5 min) and stored at -80 ℃ as virus stock for further analyses.
-
To detect SFTSV RNA, total RNA was purified from the supernatants of cell culture using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RT-PCR was performed using primers specific to the S, M, and L segments. To detect viral protein expression in Vero cells, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% TritonX-100. After three washes, cells were blocked with 5% bovine serum albumin (BSA) at 37 ℃ for 2 h. The cells were then incubated with the primary antibody (polyclonal antibodies against SFTSV nucleoprotein [NP], 1:1000 diluted in phosphate-buffered saline [PBS] with 1% FBS) at 4 ℃ overnight, followed by incubation with the secondary antibody (goat anti-rabbit serum conjugated with fluorescein isothiocyanate [FITC]; Abcam, UK). The green fluorescence was visualized using a fluorescence microscope (ECLIPSE TE2000-S; Nikon, Japan).
-
Virus particles in the supernatants were visualized by negative staining EM analysis. Supernatants from each sample (1 mL) were centrifuged at 3000 × g for 20 min at 4 ℃ to remove cell debris. The clarified supernatants were then centrifuged at 13000 × g for 40 min at 4 ℃. The pellets were resuspended in 10 μL PBS. Formvar carbon-coated copper grids were floated in droplets of virus suspension for 10 min and stained with 2% phosphotungstic acid for 1 min at room temperature. Subsequently, the grids were examined by transmission electronic microscopy.
-
The viral growth properties of SFTSV isolates in vitro were characterized by one-step growth curve analysis. Vero cells (5 × 105) were infected with viruses at a multiplicity of infection (MOI) of 10 TCID50 units/cell. Fifty microliters of supernatant was harvested from each sample at the indicated time points. Virus titers were measured by end-point dilution assays as previously described (Yu et al., 2011). Significant differences (P values) were analyzed by one-way analysis of variance (ANOVA) (Hewson et al., 2004).
-
Complete sequencing of S, M, and L segments of SFTSV isolates was performed using primers listed in Supplementary Table S1. Phylogenetic analysis was conducted based on the S, M, and L segments of SFTSV. Sequences were aligned using CLUSTALW. Phylogenetic trees were built by Mega 6.0 using maximum likelihood (ML) methods and tested by the bootstrap method with 500 replicates.
Virus isolation from serum samples of SFTS patients
RT-PCR and immunofluorescence assays (IFAs)
Negative staining electron microscope (EM)
One-step growth curve analyses
Complete genome sequencing and phylogenetic analysis of SFTSV isolates
-
Three patients (cases A, B, and C) from Hubei Province, China, were admitted to Wuhan Union Hospital from April 2015 to June 2016. The personal information, clinical features, and results of laboratory tests were summarized in Table 1. All three patients were farmers over 50 years old; two were from Macheng City (cases A and C) and one was from Guangshui City (case B). Case B reported that he was bitten by an unknown wild insect 2 weeks before the onset of disease. All patients had fever, headache, fatigue, myalgia, nausea, vomiting, and diarrhea when they presented in the hospital 4-5 days after illness onset. They had leukocytopenia and thrombocytopenia. Moreover, severe proteinuria (2 + to 3 +) and hematuria (3 +) was observed, and serum biochemistry tests revealed elevated levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), lactate dehydrogenase (LDH), and creatine kinase (CK). Because patients came from epidemic areas of SFTS and presented typical symptoms and abnormal laboratory results, they were suspected to have been infected by SFTSV. SFTSV RNA was further detected by qRT-PCR from serum samples (Table 1), confirming the occurrence of SFTS in these patients. After treatment, cases A and C recovered and were discharged on the 21th day post illness onset. However, the condition of case B worsened as he developed more severe neurologic symptoms (blurred mind and hyperspasmia), respiratory symptoms (rales in lung and dyspnea), and hemorrhagic manifestations (bleeding in mouth and impaired coagulation). He was transferred to the intensive care unit (ICU) three days after admission. Because his family decided to quit therapy, case B was discharged after spending one day in ICU. Case B died after 10 days post illness onset.
Case A Case B Case C Personal information Age and gender 52, female 59, male 67, male Occupation Farmer Farmer Farmer Admission day May 13, 2015 (the 4th day after illness onset) June 21, 2015 (the 5th day after illness onset) April 7, 2016 (the 5th day after illness onset) Location Macheng Guangshui Macheng Outcome Survived Fatal Survived Clinical manifestation on admission Fever Headache Fatigue Myalgia Yes♮ Yes Yes Yes Yes (38 ℃) Yes Yes Yes Yes (39.5 ℃) Yes Yes Yes Gastrointestinal symptoms Diarrhea Diarrhea Abdominal pain Nausea Vomiting Diarrhea Abdominal pain Respirotory symptoms Rales in lung Rales in lung Dyspnea Rales in lung Neurologic symptoms Headache Headache Blurred mind Limb tremor Headache Blurred mind Blood counts WBC (× 109/L) 3.37↓ 1.97↓ 2.16↓ PLT (× 109/L) 40↓ 16↓ 21↓ Urine routine test Proteinuria +++ ++ +++ Hematuria +++ +++ +++ Serum biochemistry ALT (U/L) 75↑ 91↑ 78↑ AST (U/L) 353↑ 291↑ 291↑ LDH (U/L) 4309↑ 4602↑ 192 CK (U/L) 1502↑ 1096↑ 666↑ CK-MB (U/L) 9.5 3.7 0.9 Laboratory tests qRT-PCR (TCID50/mL) 9.74 × 104 3.41 × 105 4.99 × 105 Virus isolation HBMC16 HBGS13 HBMC5 Note: WBC, white blood cells; PLT, platelet count; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CK, creatine kinase; LDH, lactate dehydrogenase. CK-MB, creatine kinase isoenzyme. aThe accurate body temperature of the patient is uncertain. Clinical parameters out of the normal range are shown in bold numbers with up or down arrows. Table 1. Personal information, clinical features, and laboratory tests for SFTS patients
-
Serum samples collected from the three SFTS patients collected on the first day of admission were incubated with Vero cells. After three passages, the supernatants were harvested. RT-PCR was performed, detecting S, M, and L segments in all three samples (data not shown). Blastn comparisons showed that the sequences of the PCR products had very high similarity (95%-99%) to SFTSV strains from Henan Province (data not shown). Furthermore, SFTSV NP expression was detected in Vero cells by IFA (Figure 1A). Virus particles were visualized in supernatants of each sample by negative staining EM analysis, presenting a typical morphology of bunyavirus as enveloped spherical particles with an average diameter of~100 nm (Figure 1B). Taken together, these results identified new SFTSV strains isolated from serum samples of three patients with SFTS; these strains were designated as HBMC16_human_2015 (case A), HBGS13_human_2015 (case B), and HBMC5_human_ 2016 (case C).
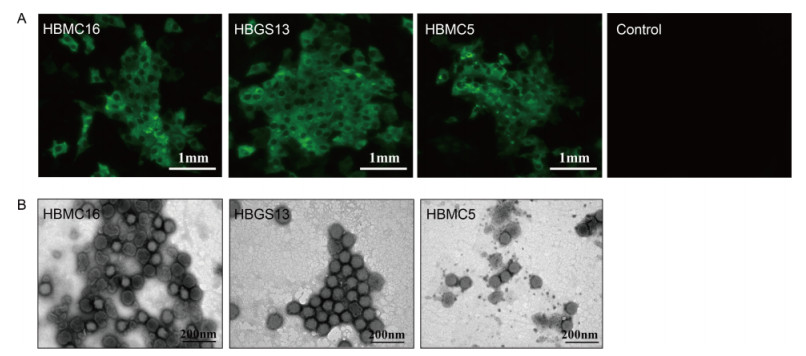
Figure 1. Isolation of the three new SFTSV strains in cell culture. (A) Detection of viral protein expression in Vero cells. SFTSV NP expression in Vero cells was detected by immunostaining using IFA after three generations of blind passaging. Cells showing green fluorescence were infected by the three new SFTSV strains. Bars, 1 mm. (B) Visualization of virus particles in cell culture supernatants by negative staining EM analysis. Bars, 200 nm.
One-step growth curve analysis was performed to characterize SFTSV growth properties in vitro. The three isolates showed similar growth properties, with yields of progeny viruses increasing rapidly beginning at 12 h post infection (p.i.) and reached a plateau at 72 h p.i. (P > 0.05; Figure 2).
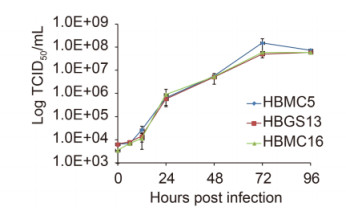
Figure 2. One-step growth curves of three new SFTSV strains.Vero cells were inoculated with an MOI of 10 TCID 50 units/cell. Supernatants were harvested at 6, 12, 24, 48, 72, and 96 h p.i. Virus titers were determined by end-point dilution assays. The tests were performed in triplicate. Bars, standard deviations.
-
The complete sequences of HBMC5 (accession numbers: KY440769, KY440770, and KY440771), HBMC16 (accession numbers: KY440775, KY440776, and KY440 777), and HBGS13 (accession numbers: KY440772, KY440773, and KY440774) were sequenced and deposited in GenBank. All reported SFTSV genomes from GenBank can be classified into five genotypes: the C1, C2, C3, C4 and J clades (Shi et al., 2016). Phylogenetic trees based on the complete sequences of S, M, and L segments showed that HBMC5 and HBMC16 together with other strains from Hubei Province (HB29, HB154, HB155, and HB156) clustered into the C3 clade and were most closely related to two strains from Henan Province (HNXY_212 and HNXY_327) (Figure 3). In contrast, HBGS13 belonged to the C2 clade and clustered with other strains from Henan Province (Figure 3).
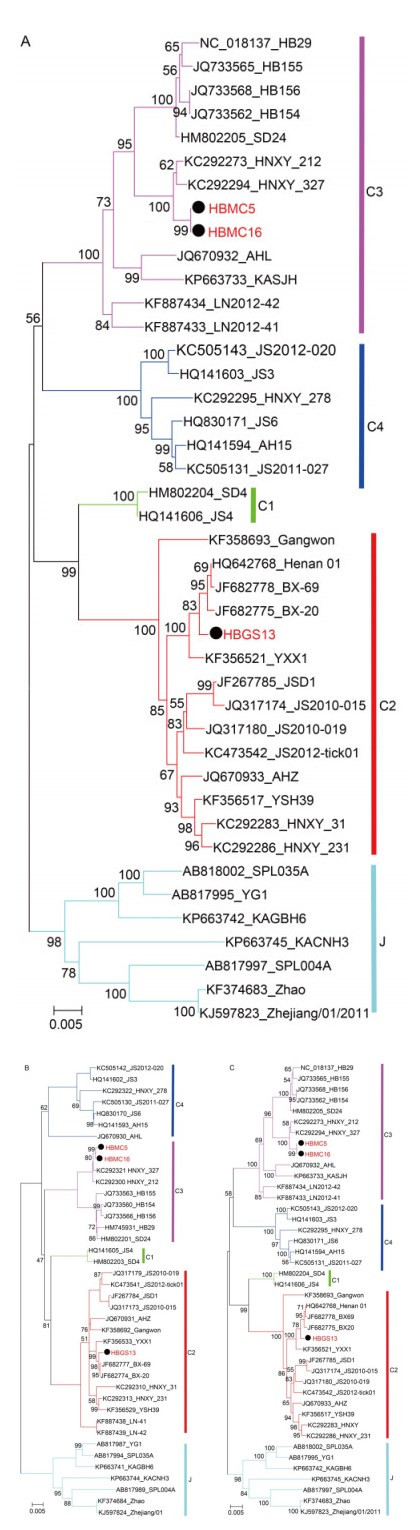
Figure 3. Phylogenetic analysis of the three new strains.The ML trees were constructed based on the complete sequences of S (A), M (B), and L (C) segments. The three isolates in this study are labeled with black solid circles. The isolates from Hubei Province are highlighted in red solid characters.
Diagnosis of SFTSV infection in patients with typical symptoms of SFTS
Isolation of novel SFTSV strains from serum samples of the three patients
Phylogenetic analysis of the three strains
-
SFTSV phylogeny is significantly associated with geographic distribution. To date, four SFTSV strains (HB29, HB154, HB155, and HB156) derived from Hubei Province have all been shown to belong to the same genotype (the C3 clade) (Shi et al., 2016). However, as limited sequences from Hubei Province were included in the previous studies (Yoshikawa et al., 2015; Shi et al., 2016), other genotypes of SFTSV strains circulating in Hubei may be concealed. Thus, obtaining more SFTSV isolates and sequences would not only provide precious genetic resources for further studies but also improve our understanding of SFTSV evolution in association with geographic distribution.
In this study, three new SFTSV strains were isolated from serum samples of SFTS patients from the Huaiyangshan area in Hubei Province, including two (HBMC16 and HBMC5) from Macheng in Huanggang City and one (HBGS13) from Guangshui in Suizhou City. Macheng and Guangshui in Hubei Province are adjacent to Xinyang City in Henan Province, where hundreds of SFTSV isolates belonging to three different genotypes (C2-C4 clades) have been identified (Xu et al., 2014; Shi et al., 2016). Phylogenetic analysis, including the complete sequences of the three strains, showed that HBMC16 and HBMC5, clustering with other Hubei isolates (HB29, HB154, HB155, and HB156), belonged to the C3 clade as well, whereas HBGS13 clustering with other isolates from Xinyang City in Henan Province belonged to the C2 clade (Figure 3). This was the first C2 genotype reported from Hubei Province, suggesting that at least two genotypes of SFTSV have been circulating in Hubei. Furthermore, in order to unveil the reality of SFTSV genotype circulation in Hubei Province, it is important to identify and isolate additional SFTSV strains from patients with SFTS; these data will provide important insights into the SFTSV phylogeny in Hubei Province and virus spreading within the epidemic areas of China.
Recently, a score model has identified that the neurologic symptoms, respiratory symptoms, viral load, and monocyte percentage were the critical risk factors for predicting the mortality of SFTS patients (Xiong et al., 2016). The clinical features of three SFTS patients were reviewed in this study, showing that case B developed severe neurologic and respiratory symptoms, and died of multiple organ failure (MOF). Because the C2 strain (HBGS13) was isolated from case B, the mortality of SFTS patients might be associated with the virus genotypes, which, however, needs further investigation. We determined the in vitro growth properties of the three isolated strains and found that there were no significant differences in progeny virus yields. So there might not be any significant differences in viral replication and proliferation of HBMC16, HBGS13, and HBMC5 although they belonged to different genotypes. Whether these strains hold similar properties in immune response and drug resistance will be tested in the future. Furthermore, identification and isolation of additional SFTSV strains will promote the understanding of the relation between virus genotypes and mortality of SFTS patients.
In summary, three new SFTSV strains were isolated from cities in the Huaiyangshan area of Hubei Province. Phylogenetic analysis found that they belonged to different genotypes, suggesting that more than one genotype of SFTSV had been circulating in Hubei. These results provided insight into SFTSV evolution and improved our understanding of SFTSV prevalence.
-
This work was supported by the Science and Technology Basic Work Program (2013FY113500) and the National Key Research and Development Program (2016YFE 0113500) from the Ministry of Science and Technology of China, as well as the European Union's Horizon 2020 EVAg project (No 653316).
-
The authors declare that they have no conflict of interest. The study was approved by both ethics committees of Wuhan Institute of Virology, Chinese Academy of Sciences (Approval number: WIVH332011601), and the Union Hospital, Tongji Medical College, HuaZhong University of Science and Technology (Approval number: 2015LSZ[031]). All participants provided written consent for anonymous use of their specimens and clinical information for research.
-
ZHH, XZ, FD, and SS conceived of the study. YFZ and SS isolated and identified the SFTSV and wrote the manuscript. XZ, CP, MYL, WJZ, and MML collected the serum samples and patient information and performed diagnostic testing and treatment. ZYS obtained the full-length SFTSV sequences. JMS generated the sequence datasets for phylogenetic analysis and constructed the phylogenetic trees. ZHH, XZ, and FD checked and finalized the manuscript.
Supplementary Table S1 is available on the websites of Virologica Sinica: www.virosin.org; link.springer.com/journal/12250.
-
Primers Sequences (5'-3') Locations of primers S segment S1F acacaaagacccccttcatttrg 1-23 S1R ACACAAAGAACCCCCAAAAAAGGA 1722-1744 M segment M1F acacaragacggccaacaatga 1-22 M1R TCTCCCAGTTGTGAYGCATTCCTTC 1767-1791 M2F GGCAACCAWGATGATGTTAGGAT 1597-1619 M2R ACACAAAGACCGGCCAACACT 3358-3378 L segment L1 F acacaragacgcccagatgrac 1-22 L1 R GAGACCACTGRACCACATTRCTG 1549-1571 L2F GTGTCAATCTTGTTGGAAAARGCAT 1463-1487 L2R GAGCTTYGAGACGAAATARGAC 3313-3314 L3F GGTTGAAGTCAGCCCGYAGTCT 2913-2934 L3R ATTCTRACTACTTGGCTTATGGTGG 4800-4824 L4-F TTAGGGAGAGAAACATTGTCAGGAG 4485-4509 L4-R acacaaagaccgcccagatctta 6346-6368 Table S1. Primers used to sequence the complete genome of SFTSV isolates
















 DownLoad:
DownLoad: