HTML
-
Porcine circovirus type 3 (PCV3) is a new-emerging circovirus belonging to the genus Circovirus in the family Circoviridae in which PCV type 1 (PCV1) and PCV type 2 (PCV2) were well documented (Palinski et al. 2017). PCV1 is a cell culture-derived virus and is considered to be nonpathogenic for swine, whereas PCV2 is the primary etiological agent of porcine circovirus-associated diseases (PCVAD) that causes severe economic losses in the swine industry worldwide (Zhai et al. 2014). PCV3 was firstly reported in the United States of America in 2016 (Palinski et al. 2017). After that, PCV3 has been detected in Poland, South Korea, Italy, Brazil, United Kingdom, and China, and was reported to be associated with porcine dermatitis and nephropathy syndrome, congenital tremors, reproductive failure, and multi-systemic inflammation (Kwon et al. 2017; Li and Tian 2017; Palinski et al. 2017; Stadejek et al. 2017; Zheng et al. 2017). The first case of PCV3 in China was reported in Guangdong Province in March 2017 (Shen et al. 2017). After that, PCV3 cases were frequently reported in other provinces in South China such as Guangxi, Jiangxi, Fujian, and Hunan provinces (Fu et al. 2017). The status of PCV3 prevalence in Central and North China was rarely reported. To figure out current status of PCV3 prevalence, a national epidemiological survey on PCV3 was conducted in this study.
A total of 730 clinical tissue samples were collected from 60 pig farms in 17 provinces and 3 autonomous municipalities with different geographical distributions (South China: Fujian, Jiangsu, Sichuan, Zhejiang, Anhui, Hubei, Jiangxi, Guangdong and Shanghai; Central China: Shandong, Shanxi, Henan, Shaanxi, and Gansu; North China: Hebei, Jilin, Inner Mongolia, Beijing, Tianjin, and Liaoning) in China. The collected samples included lung (84), lymph node (75), tonsil (58), kidney (65), brain (56), spleen (82), heart (77), liver (34), and stillbirth (199). These samples were individually collected, stored and transported in ice boxes. All animal studies in this study were approved by the Animal Care and Ethics Committee of National Research Center for Veterinary Medicine with IACUC number 2018054. The tissue samples were homogenized and diluted with phosphate-buffered saline (PBS: 0.1 mol/L, pH 7.4) and then subjected to centrifuge at 10, 000 rpm for 10 min at 4 ℃. The supernatants were then utilized for DNA extraction (Tiangen, Beijing, China) following the manufacturer's instructions. PCV3 DNA detection using conventional PCR was performed as previously described with forward primer 5'-CCACAGAAGGCGCTATGTC-3' and reverse primer 5'-CCGCATAAGGGTCGTCTTG-3' (Palinski et al. 2017). The PCR reaction was performed as follows: 94 ℃ for 3 min, followed by 35 cycles of 94 ℃ for 20 s, 55 ℃ for 30 s, 72 ℃ for 30 s, and 72 ℃ for 5 min.
One hundred and four out of 730 clinical samples were tested to be PCV3 positive using PCR and the positive rate was 14.25% (Table 1). As for the geographical distribution, the positive rates of PCV3 in South China (16.16% ± 6.04%) was significantly higher than North China (5.96% ± 3.84%) analyzed by One-way ANOVA using Sigmaplot 11 software (Systat Software Inc., San Jose, CA) (P = 0.0098). There was no statistically significant difference between South China and Central China (13.05% ± 4.62%). Since the previously published literatures about the PCV3 epidemiological survey were mainly focused on provinces in South China, the results in this study for the first time indicated that PCV3 has widely spread to central and north parts of China and has nationwide distribution.

Table 1. Sample information and positive rates in different provinces or municipalities.
PCV3 has been reported to mainly infect lymphoid tissues, such as lung, lymph nodes, and tonsil (Fan et al. 2017; Fu et al. 2017). In this study, the primary PCV3-postive tissues were found to be lung, followed by lymph nodes, stillbirth, kidney, brain, tonsil, spleen, heart, and liver (Table 2). To quantity virus genome copy numbers in above different types of tissues, a commercial real-time PCR kit (Beijing Anheal Laboratories Co., Ltd. Beijing, China) was applied. Real-time PCR was performed using an ABI 7500 instrument (Applied Biosystems, Foster City, USA). The real-time PCR mixture contained 12.5 lL Premix Ex Taq (Takara, Dalian, China), 1 lL template DNA, 1 lL primers (1 lmol/L), 1 lL probe (0.5 lmol/L), and sterile water to bring the final volume to 25 lL. The amplification parameters were set as 95 ℃ for 30 s, followed by 40 cycles of 95 ℃ for 10 s, and 60 ℃ for 30 s. The means of Ct values of different types of tissues were used to reflect the relative amount of virus genome copy number according to the product manufacture's instruction. The highest genome copy numbers were found in kidney samples, followed by stillbirth, lung, lymph nodes, tonsil, brain, spleen, heart, and liver (Table 2). The mean Ct values of PCV3-negative samples were 38.7 ± 2.3.

Table 2. Percentage of PCV3-positve tissues and Ct values of realtime PCR.
To figure out potential multiple infections of PCV3-postive tissue samples, PCV3-postive samples were also used to detect other porcine viruses including PCV2, porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), porcine parvovirus (PPV), and classical swine fever virus (CSFV) using conventional PCR as previously described (Li et al. 2013, 2014, 2017; Lv et al. 2016). Consistent with previous reports (Fu et al. 2017), PCV2 and PRRSV were the primary viral pathogens in multiple infections. Thirty-five, 41, and 26 PCV3-positive clinical samples were found to be co-infected with PCV2, PRRSV, and PCV2/PRRSV, respectively. No co-infection with CSFV, PRV, and PPV was observed in this study. There were only two cases of singular PCV3 infection without other viruses detected in above PCV3-positive clinical samples. The above results were consisted with previous reports in which co-infections of PCV3 with other swine pathogens such as PCV2, PPRSV, and astrovirus were frequently detected and may aggravate the clinical signs (Phan et al. 2016; Fu et al. 2017).
To get complete PCV3 genome sequence of positive samples, two pairs of primers was used as previously described (Kwon et al. 2017). The complete genomes of six PCV3 isolates in this study were obtained and deposited in GenBank under the accession numbers from MF769806 to MF769811. To perform phylogenetic analysis, PCV3 genome sequences obtained in this study and from NCBI were aligned using Clustal W, and a phylogenetic tree was constructed using the maximum-likelihood method with 1000 bootstrap replicates in MEGA7.0 software (Kumar et al. 2016). A phylogenetic tree for full-length Cap gene was also constructed as described above.
Six PCV3 genomes were obtained from above PCV3-postive samples. Among of them, one PCV3-postive samples came from South China (Jiangsu Province), four came from Central China (Henan and Shanxi Province), and one came from North China (Beijing). The above six PCV3 genomes in this study were compared with other reported PCV3. As shown in Fig. 1A, six PCV3 isolates were clustered in different branches, and shared 98.9%–100% genome similarity with other reported strains. There were no distinct branches formed among all reported PCV3s according to geographical distribution and time of virus isolation. In Fu's study, PCV3 isolates were divided into PCV3a, PCV3b, and PCV3c clades according to two amino acid mutations (A24 V and R27 K) on Cap gene (Fu et al. 2017). The full-length of six PCV3 Cap genes were also analyzed by above amino acids and compared with other reported PCV3 strains. As shown in Fig. 1B, two PCV3 strains from Henan (Central China) were clustered into clade PCV3a, two PCV3 strains from Beijing (North China) and Jiangsu (South China) were clustered into clade PCV3c, and two PCV3 strains from Shanxi and Henan (Central geographically spread to Central and North China (Table 1).
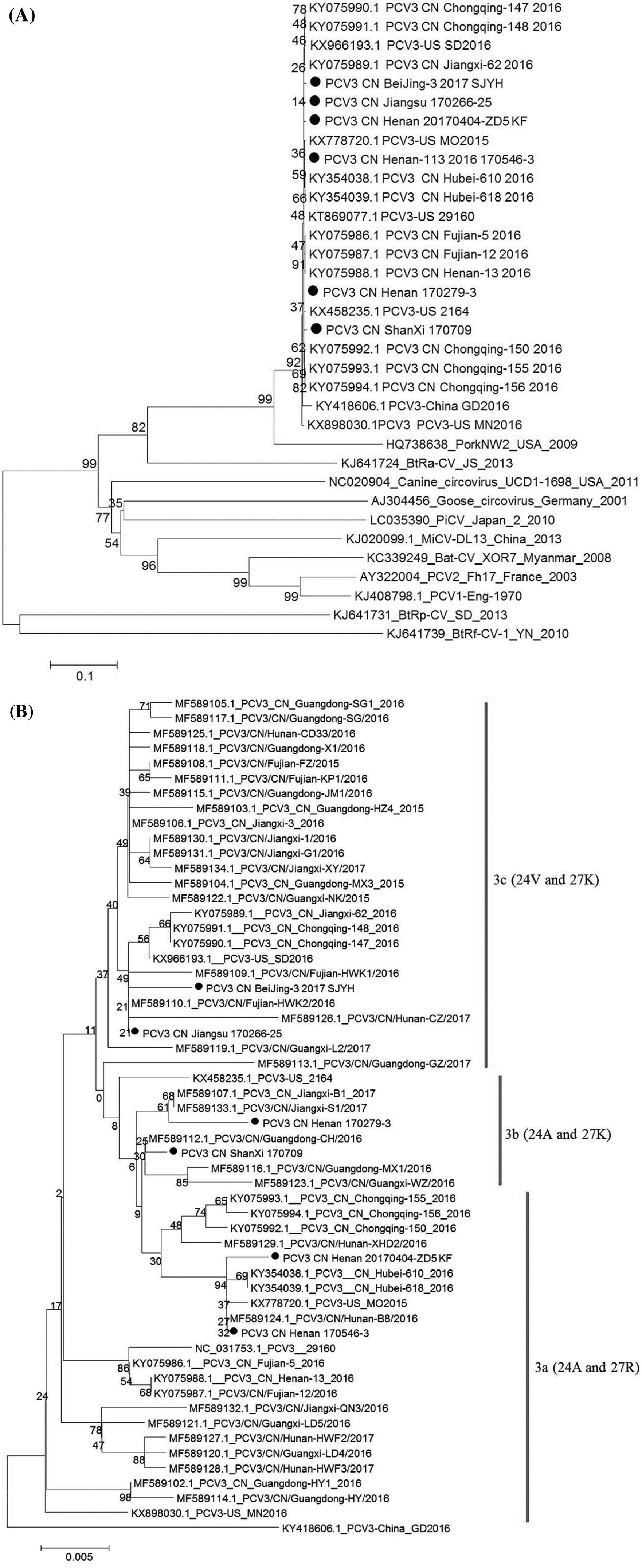
Figure 1. Phylogenetic analysis of PCV3 strains. A Whole genomebased phylogenetic relationship of six PCV3 isolates in this study with other reported PCV3. B Full-length Cap gene-based phylogenetic relationship of six PCV3 isolates in this study with other reported PCV3. Dot indicates the PCV3 isolates in this study.
So far, the pathogenicity of PCV3 has not been evaluated due to the failure of PCV3 isolation on cells. Also, singular PCV3 infection was rarely detected in clinical samples and several severe pathogenic porcine viruses such as PRRSV and PCV2 were found to be co-infected with PCV3. Therefore, studies about the pathogenicity of singular PCV3 infection and/or PCV3 co-infected with other important viral pathogens on pigs should be the priority to elucidate the significance of PCV3 on swine industry.
-
This work was supported by grant from National Key Research and Development Program (2016YFD0500703), Major science and technology projects in Henan Province (171100110200), and Luoyang HeLuo Talent Plan (to Dr. Kegong Tian).
Acknowledgements
-
The authors declare that they do not have any conflict of interest.
-
Statement This study was approved by the Animal Care and Ethics Committee of National Research Center for Veterinary Medicine with IACUC Number 2018054.














 DownLoad:
DownLoad: