HTML
-
Ascoviruses, belonging to the Ascoviridae family of insectspecific pathogens, contain a large circular double-stranded DNA (dsDNA) genome of 100–200 kb (Asgari et al. 2017), assembled in allantoid or rod-like virions (Federici 1983; Bigot et al. 1997; Cheng et al. 1999; Huang et al. 2012b; Wei et al. 2014; Huang et al. 2017; Arai et al. 2018). The size of the virions ranges from 200 to 400 nm. They are embedded in vesicles and accumulate in the host larval hemolymph (Federici et al. 1990; Federici and Govindarajan 1990). Ascoviruses can be divided into five major species: Spodoptera frugiperda ascovirus 1 (including SfAV-1a and SfAV-1d), Trichoplusia ni ascovirus 2 (including TnAV-2a and TnAV-2b), Heliothis virescens ascovirus 3 (including HvAV-3a, HvAV-3e, HvAV-3h, HvAV-3i, and HvAV-3j), Trichoplusia ni ascovirus 6 (including TnAV-6a and TnAV-6b, previously named as TnAV-2c and TnAV-2d respectively), and Diadromus pulchellus toursvirus 1 (including DpTV-1a, previously named as DpAV-4a) (Carner and Hudson 1983; Hamm et al. 1986; Cheng et al. 2000; Asgari et al. 2007; Cheng et al. 2005; Huang et al. 2012b; Asgari et al. 2017; Huang et al. 2017; Arai et al. 2018; Liu et al. 2018; Chen et al. 2018, 2019). Heliothis virescens ascovirus 3j (HvAV-3j) is a recently reported isolate of Heliothis virescens ascovirus 3a from Spodoptera litura in Japan, and can be transmitted by the endoparasitoid Meteorus pulchricornis under laboratory conditions (Arai et al. 2018).
Laboratory data indicated that ascoviruses have the potential to develop into biopesticide owing to their high virulence to noctuid pests, such as Spodoptera exigua, S. litura, and Helicoverpa armigera (Federici 1982, 2009; Li et al. 2013, 2016; Yu et al. 2018b). Unlike the welldeveloped baculoviruses, the poor per os infectivity and the parasitoid-dependent transmission are the main possible constraints to the employment of ascoviruses for the development of bioinsecticides. In addition to their use as bioinsecticides, baculoviruses are widely employed as eukaryotic expression vectors and gene therapy vectors (Volkman and Goldsmith 1983; Carbonell et al. 1985), also due to their low environmental impact and lack of infectivity to non-target organisms (Arif 2005). As for ascoviruses, they have been only isolated from insect hosts, but infectivity to mammalian cells is not well documented.
In order to explore the infectivity of ascoviruses toward mammals, accurate in vitro detection was performed in four insect (Sf9, HzAM1, SeFB, and HaFB) and six mammalian cell lines (HEK293, 7402, HePG2, PK15, ST, and TM3), and the results were compared to those obtained using a baculovirus. This study provides insights into the evaluation of ascovirus-related risks to non-target organisms.
-
Four insect and six mammalian cell lines were used to evaluate the infectivity of HvAV-3j. SeFB (IOZCAS-Spex-Ⅱ-A) (Zhang et al. 2009) and HaFB cells were derived from the fat bodies of S. exigua and H. armigera, respectively. Sf9 and HzAM1 cells were obtained from ovary of S. frugiperda and H. zea, respectively. These four different insect cells were maintained in TMN-FH insect culture medium (Sigma, USA) with 10% FBS (GIBCO, USA) at 27 ℃. HEK293 cells were derived from human renal epithelial cells; 7402 and HePG2 cell lines were derived from human lung cancer cells; PK15, ST, and TM3 cell lines were derived from porcine kidney epithelial, pig testis, and mouse testicular mesenchymal cells, respectively. These mammalian cells were cultured in DMEM medium (Hyclone, USA) with 10% FBS (GIMINI, USA) at 37 ℃ and 5% CO2. HvAV-3j was stored at 4 ℃ in the hemolymph collected from the infected S. exigua larvae containing 0.5% phenylthiourea. A green florescence protein-encoding Autographa californica nucleopolyhedrovirus (AcMNPV-Egfp) was constructed and maintained in Sf9 cells and the budded viruses contained in the supernatant at multiplicity of infection (MOI) = 5 were used to inoculate different cultured cells for the following experiments.
-
The laboratory-maintained beet armyworm, S. exigua, was used for ascovirus infection and cultured on an artificial diet at 27 ± 1 ℃ and a 16/8 h (light/dark) photoperiod. To inoculate the ascovirus, the HvAV-3j-containing hemolymph mixture (1.1 × 1011 virion copies/mL) was prepared according to Li et al. (2016). HvAV-3j-containing S. exigua hemolymph was diluted 1000-fold with FBS-free TNM-FH medium. Diluted ascovirus-containing medium was sterilized by filtration with a 0.22-μm filter (Millipore, USA). The FBS was added to a final concentration of 10%. Hemolymph collected from S. exigua uninfected larvae was diluted and sterilized in the same way, and used as negative control (mock-infected control) in the following assays.
Cells were seeded in 6-well plates with a primary density of 105 per well in 1 mL of medium and allowed to attach for 1 h. One milliliter of prepared HvAV-3jcontaining medium, negative control medium, or AcMNPV-containing medium (105 TCID50 mL-1) were added into each well, as appropriate. To avoid repeated infection, the supernatant of each well was replaced with fresh culture medium after 1 h of infection, and this point was set as 0 h post-infection (hpi). At 0, 3, 6, 12, 24, 36, 48, 60, 72, 96, 120 and 168 hpi, the cell morphology was evaluated by reverse microscopy. Cells at the different time points post-virus inoculation were used to investigate the cell viability by the MTT method (Kim et al. 2007). To this end, cells were transferred into 96-well plates and, after allowing them to attach for 1 h, viruses were added. At different time points post-virus infection, the supernatant of each well was removed and 100 μL of 1 mg/mL 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) was added into each well. After a 4-h incubation, the culture medium in each well was replaced with 150 μL of DMSO. The absorbance at 490 nm was measured, and relative cell viability calculated as described by Kim et al. (2007).
At each time point, the differences in cell viability between mock-infected, HvAV-3j-infected, and AcMNPVEgfp-infected cells were analyzed by two-way analysis of variance (ANOVA) using a least significant difference (LSD) test via SPSS (v16, SPSS Inc, Chicago, IL, USA). The general linear method (GLM) in SPSS was used to evaluate the differences in cell number and viability between the three treatments.
-
Cells seeded in 6-well plates as above described were infected with HvAV-3j or AcMNPV-Egfp. At 0, 3, 6, 12, 24, 48, 72, 96, and 120 hpi, the cells were collected for DNA, RNA, and protein extraction. The DNA was extracted by using the TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 (TaKaRa, JPN), according to the manufacturer's instructions. The total RNA was extracted using TRI reagent (MRC, USA), according to the manufacturer's instructions, and the firststrand cDNA was obtained by reverse transcription with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) using 1.0 μg of total RNA as a template. The chloroform phase of each sample during the RNA extraction was used to obtain the total protein, according to the manufacturer's instructions for TRI reagent. For each time point, three replicate extractions of DNA, RNA, and protein samples were performed.
-
The DNA replication curve of the recombinant viruses was investigated by qPCR as described by Li et al. (2016). The sequences of primers used for qPCR detection are shown in Supplementary Table S1. The GLM in SPSS 15.0 (SPSS Inc, Chicago, IL) was used to compare the differences in DNA replication between the different virus strains.
-
The cDNA synthesized from different cellular total RNAs, as described above, was used as template for PCR-based detection with the gene-specific primers shown in Supplementary Table S1. The major capsid protein gene (mcp, orf56) encodes the structural protein of HvAV-3j; the DNA polymerase (Dpol, orf1) and helicase genes (helicase, orf133) are genomic DNA synthesis-associated genes of HvAV-3j; the inhibitor of apoptosis protein 1 gene (iap1, orf22) and caspase-like gene (casp, orf176) are apoptosisassociated viral genes; the glycosyl transferase gene (GTase, orf87) is involved in carbohydrate metabolism. For AcMNPV detection, the Dpol (orf65), 38 K (orf98), gp64 (orf128), and polyhedrin (polh, orf8) were selected as the representatives of different functional genes. Gapdh was used as reference gene.
-
Total proteins extracted from the different HvAV-3jinfected cells were analyzed by western blotting to evaluate the expression of the major capsid protein (MCP). The protein samples were separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane. A polyclonal antibody against HvAV-3h MCP (1:3000; Yu et al. unpublished) and a monoclonal anti-b-actin antibody (1:4000; Proteintech, Cat No. 66005-1, CHN) were used as primary antibodies. The noctuid insect-specific GAPDH antibody (1:4000) was used as reference antibody (Yu et al. 2018a). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit (1:5000) or HRP-conjugated goat anti-mouse μgG (1:5000) (Proteintech, CHN), were used as the secondary antibodies. The proteins were visualized by using ClarityTM Western ECL Substrate (Bio-Rad, USA).
Cells and Viruses
Cell Infection and Cell Viability Assays
Extraction of DNA, RNA, and Protein
Detection of Viral Genomic DNA Replication
Detection of Viral Gene Transcription
Detection of Viral Protein Expression
-
After exposure to HvAV-3j, significant morphological changes were observed in the four different tested insect cells. Aggregation and swelling of host cells were typical alterations caused by HvAV-3j infection (Fig. 1A, panel 1). The HvAV-3j-infected Sf9 cells acquired a round shape from the first day post-infection (dpi), and started to detach at 2 dpi; aberrant vesicles composed Sf9 cells were observed upon HvAV-3j infection at 2, 3, and 5 dpi; on the fifth day, the appearance of numerous spots indicated Sf9 cells destroyed by HvAV-3j infection. Swelling and rounding were also observed in HvAV-3j-infected HzAM1 cells (Fig. 1A, panel 2). Although typical vesiclecontaining cells were also detected among infected HzAM1 cells, most of these cells were still intact at 5 dpi. SeFB cells suffered the most severe impact from HvAV-3j infection (Fig. 1A, panel 3). The mock-infected SeFB cells displayed a fibroblast-like appearance and maintained adherence to the surface of culture flasks. Upon HvAV-3j infection, above 60% of SeFB cells acquired a round shape and detached at 1 dpi; aggregated SeFB cells gradually broke into debris from 2 to 5 dpi; at 5 dpi, few swelled SeFB cells were still floating. HaFB cells were rarely destroyed by HvAV-3j infection, even at 5 dpi (Fig. 1A, panel 4). However, infected HaFB cells detached from the bottom of flasks and appeared granular from the first day post-infection. Thus, although infection by HvAV-3j induced swelling and floating in Sf9, HzAM1, SeFB, and HaFB cells, significantly different responses were observed in distinct cell lines.
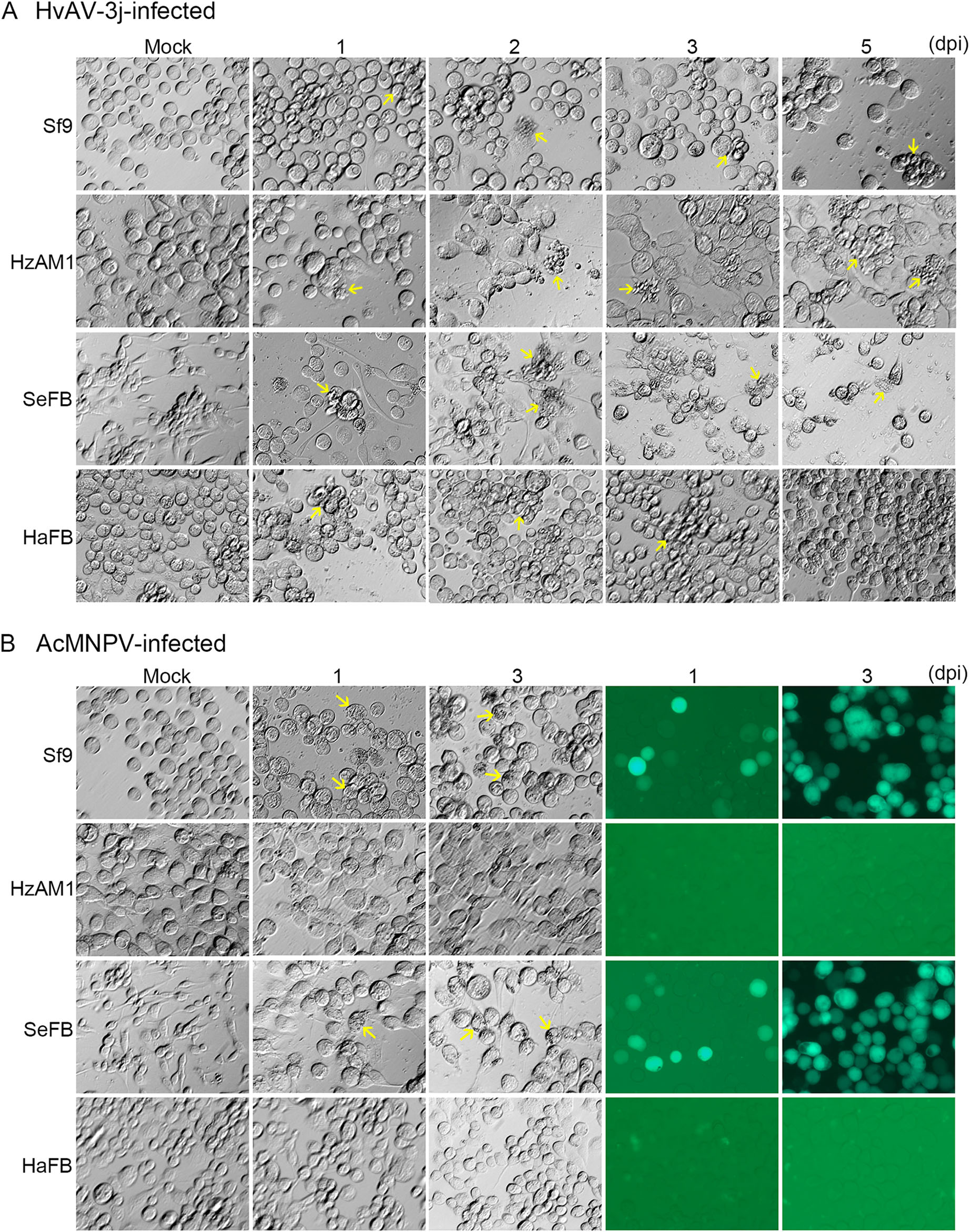
Figure 1. Microscopy analysis of virus-infected insect cell lines. Morphological changes in Sf9, HzAM1, SeFB, and HaFB cells inoculated with HvAV-3j (A) or AcMNPV-Egfp (B). Arrows in A: aggregated and malformed cells due to HvAV-3j inoculation; arrows in B: occlusion bodies induced by AcMNPV-Egfp. Magnification of all figures: 200×.
In the AcMNPV-Egfp-infected Sf9 and SeFB cells, occlusion bodies typical of baculoviruses were found, and green fluorescence could be detected from the first day post-infection (Fig. 1B, panel 1, 3). In contrast, HzAM1 and HaFB cells grew normally after exposure to AcMNPVEgfp, and no green fluorescence was detected from AcMNPV-Egfp-infected HzAM1 and HaFB cells (Fig. 1B, panel 2, 4). These results indicated that AcMNPV was infectious to Sf9 and SeFB cells, but not to HzAM1 and HaFB cells.
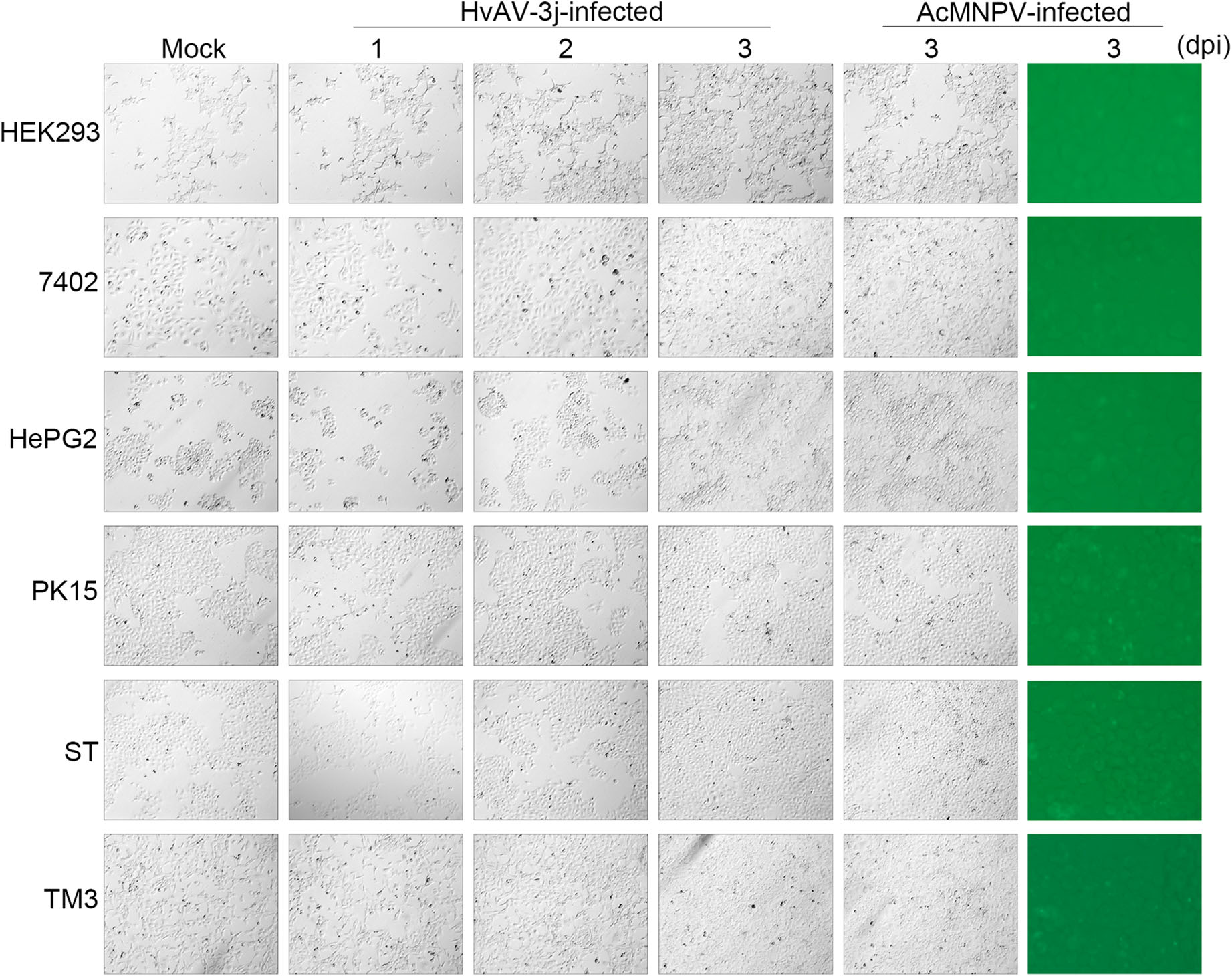
The effects of HvAV-3j and AcMNPV-Egfp infection were also explored in mammalian cells by monitoring cell morphology and EGFP expression (Fig. 2). Six mammalian cell lines were selected, i.e., three human (HEK293, 7402, HePG2) and two pig cell lines (PK15, and ST), as well as a mouse cell line (TM3). All the selected cell lines grew normally in the presence of both HvAV-3j and AcMNPV-Egfp, and no green fluorescence could be detected. These results indicated that neither HvAV-3j nor AcMNPV can infect these six mammalian cells.
-
To further characterize the impact of viral infection on cell proliferation, the relative cell viability was assessed by MTT assays. The infection by HvAV-3j nearly arrested Sf9, HzAM1, SeFB, and HaFB cell proliferation, and a significant inhibition of cell growth was observed at 12 hpi (Fig. 3). The proliferation indexes (PIs) of all HvAV-3j-infected insect cells drastically decreased from 6 to 12 hpi. In particular, PI decreased by 41.5% in Sf9 cells (t = 16, df = 8, P < 0.0001) and HzAM1 cells (t = 8.068, df = 8, P < 0.0001), by 39.8% in SeFB cells (t = 3, df = 8, P = 0.0128), and by 40.2% in HaFB cells (t = 3.253, df = 8, P = 0.0117). These results indicated that the HvAV-3j significantly inhibited the growth of host cells, and acute pathogenicity was observed between 6 and 12 hpi.
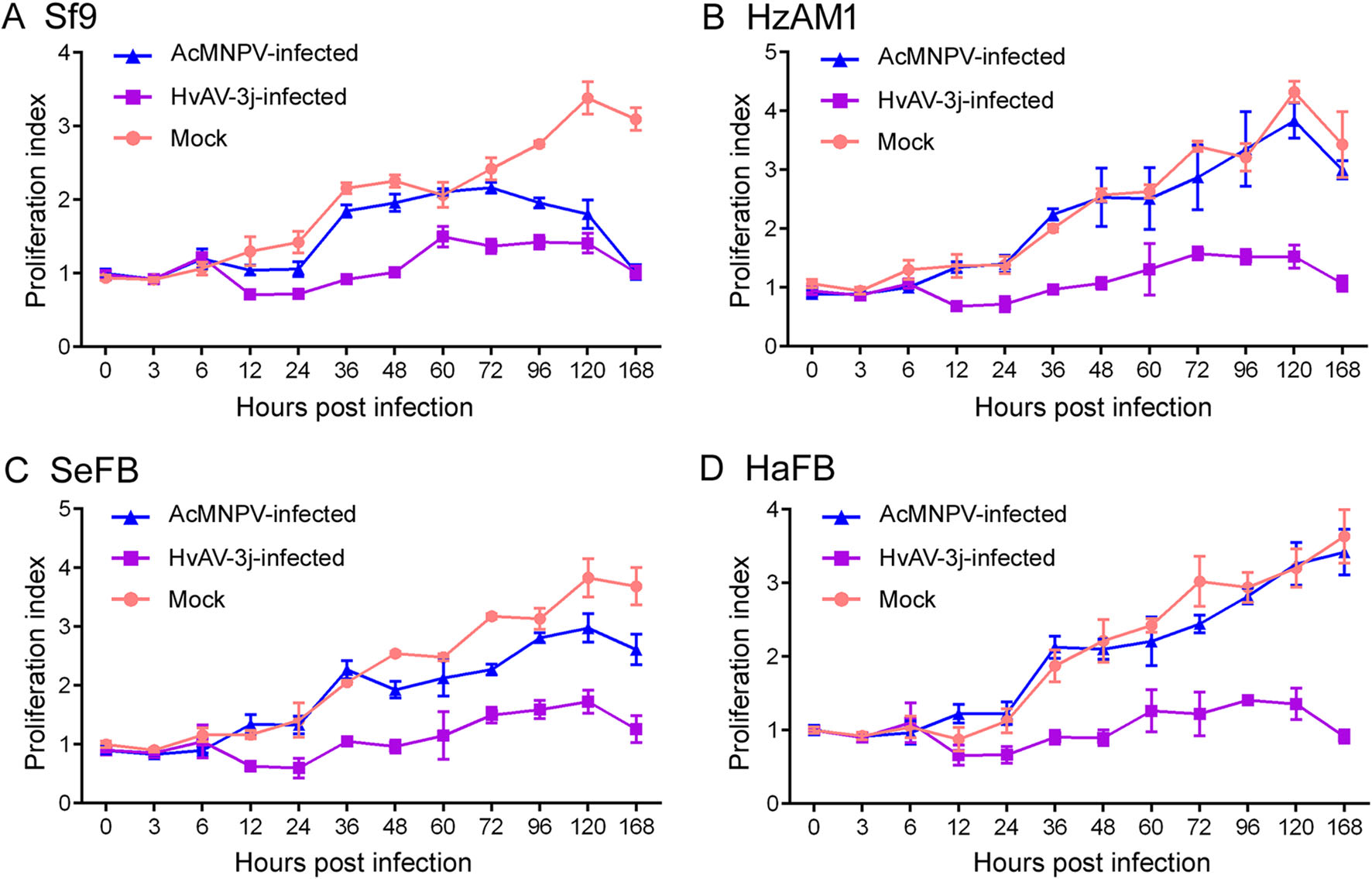
Figure 3. Insect cell viability after virus infection. Proliferation index measured by MTT assay after inoculation with HvAV-3j or AcMNPV-Egfp. A Sf9 cells. B HzAM1 cells. C SeFB cells. D HaFB cells.
The MTT assays on AcMNPV-Egfp-infected insect cells were in close agreement with the results of the above described microscopy evaluation. According to the GLM analysis, no significant differences were observed between the PI of mock-infected and AcMNPV-Egfp infected HzAM1 (F = 1.790, df = 11, 144, P = 0.082) and HaFB (F = 1.249, df = 11, 144, P = 0.283) cells. On the other hand, AcMNPV infection significantly decreased the PI of Sf9 (F = 85.28, df = 11, 144, P < 0.0001) and SeFB (F = 29.85, df = 11, 144, P < 0.0001) cells, but this index was significantly higher compared to HvAV-3j-infected Sf9 (F = 293.8, df = 11, 144, P < 0.0001) and SeFB (F = 245.6, df = 11, 144, P < 0.0001) cells. These results indicated that AcMNPV had a lower impact on host cell growth (Sf9 and SeFB) compared to HvAV-3j.
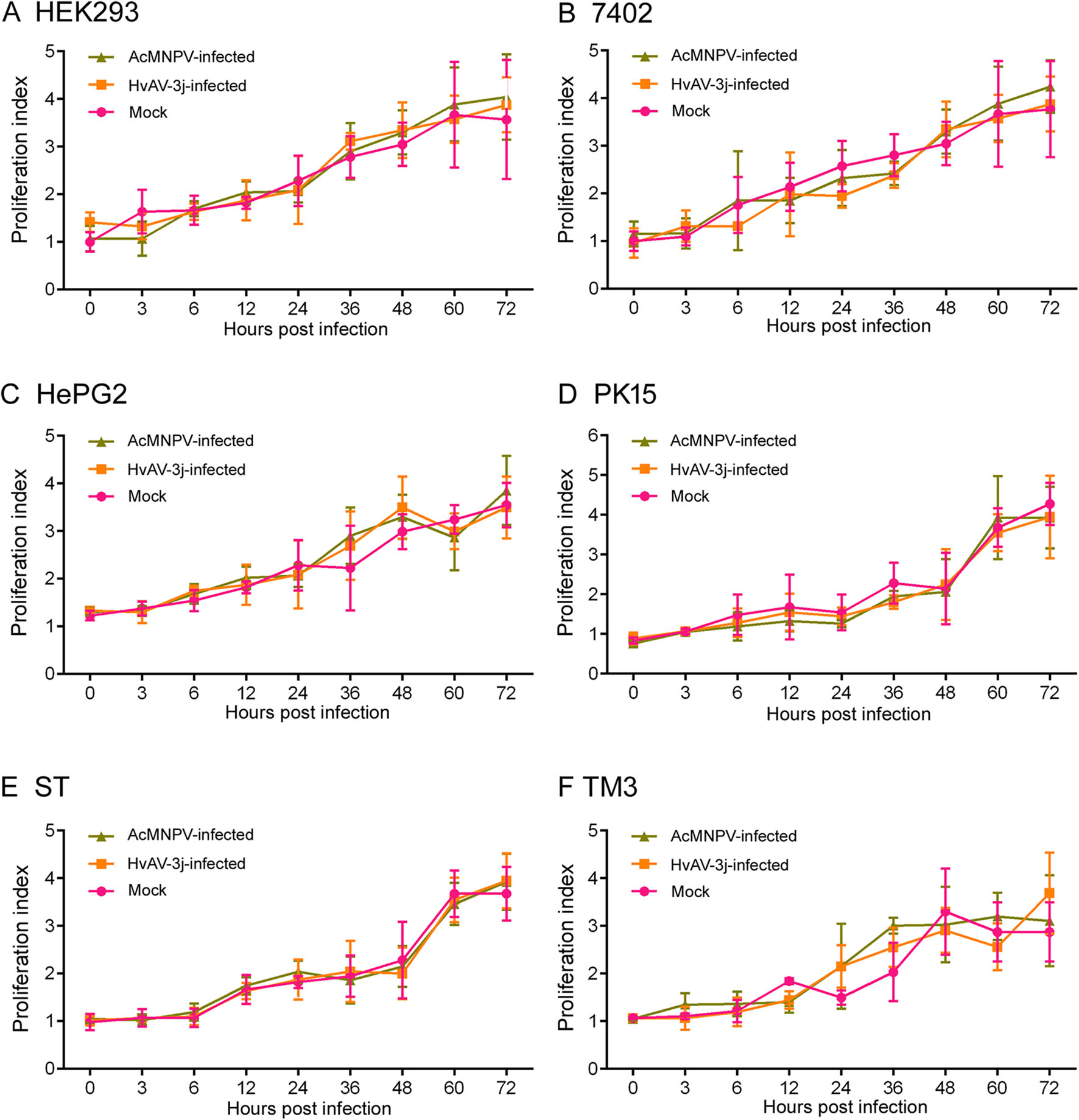
MTT assays were also performed on HEK293, 7402, HePG2, PK15, ST, and TM3 cells to investigate the infectivity of HvAV-3j or AcMNPV on mammalian cells (Fig. 4). The results showed that in mammalian cells exposed to both HvAV-3j and AcMNPV-Egfp growth was comparable to that of mock-infected cells, in accordance with the above described microscopic observations.
-
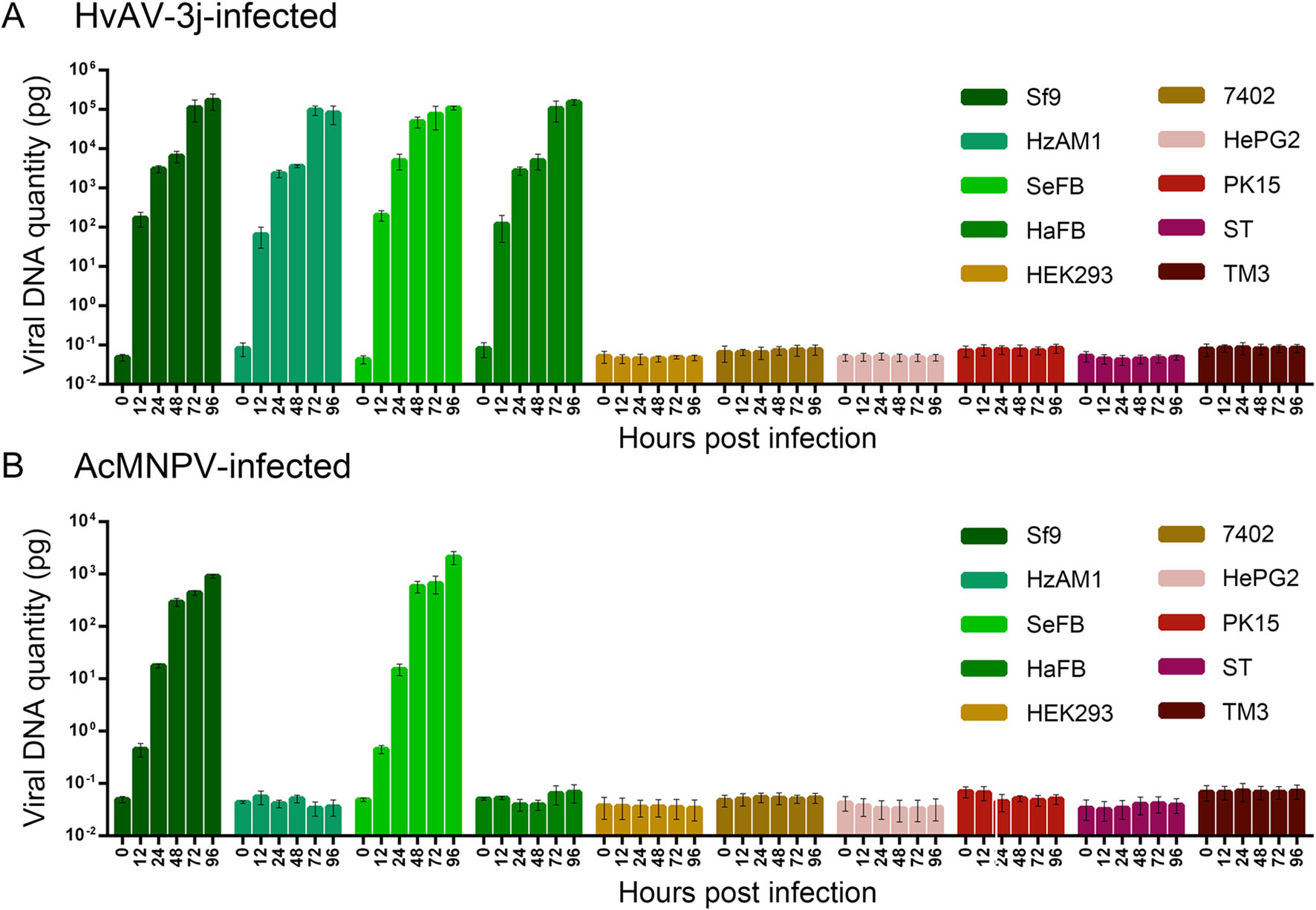
In order to examine viral replication in the different cell lines, the HvAV-3j and AcMNPV-Egfp viral DNA content was analyzed by qPCR (Fig. 5). HvAV-3j viral DNA replication was similar in infected Sf9, HzAM1, SeFB, and HaFB cells (Fig. 5A). An interesting finding was that the HvAV-3j genomic DNA content increased at least one thousand-fold in the first 12 h after infection in all four insect cell lines examined, and in SeFB cells, HvAV-3j could produce millions of genome copies within 48 h after infection. Unlike HvAV-3j, AcMNPV-Egfp could only replicate in Sf9 and SeFB cells and finally yield ten thousand copies of its genome after 96 h of infection, corresponding to only 1% of HvAV-3j replication efficiency which was only 1% of HvAV-3j replication folds change (Fig. 5B). No viral DNA replication was detected from HvAV-3j- or AcMNPV-Egfp-infected HEK293, 7402, HePG2, PK15, ST, and TM3 cells, which indicated that neither HvAV-3j nor AcMNPV were infectious to mammalian cells.
-
The transcription of viral genes was analyzed in HvAV-3j-infected insect cells to further explore viral infectivity. Among the HvAV-3j-encoded genes related to apoptosis, the genes for inhibitor of apoptosis protein 1 (iap1, orf22) and a caspase-like protein (Casp, orf176) started to be transcribed from 12 hpi. Transcription of the replicationassociated HvAV-3j genes helicase (orf133) and DNA polymerase (Dpol, orf1) started at 12 hpi and continued until 96 hpi. The HvAV-3j gene for the structural major capsid protein (mcp, orf56) started to be transcribed at 24 hpi. Finally, transcription of the HvAV-3j gene for glycosyl transferase (GTase, orf87), implicated in carbohydrate metabolism, started at 12 hpi and continued until 96 hpi (Fig. 6A).
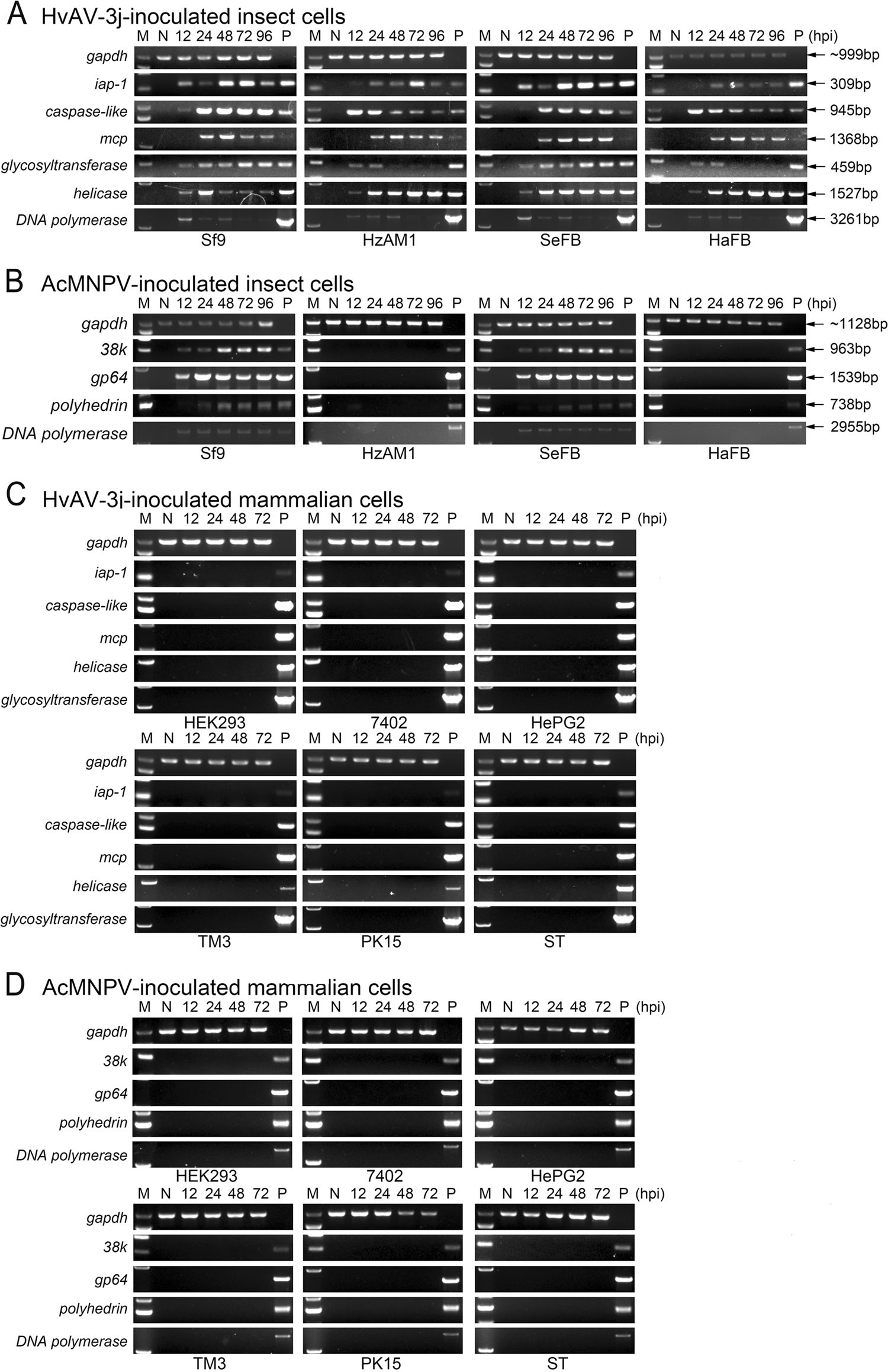
Figure 6. Transcription of selected viral genes in different cells. A Reverse transcription PCR (RT-PCR) of HvAV-3j-encoded iap-1, caspase-like, mcp, glycosyltransferase, helicase, and DNA polymerase in HvAV-3j-inoculated insect cells. B RTPCR of AcMNPV-encoded 38k, gp64, polyhedrin, and DNA polymerase in AcMNPV-Egfpinoculated insect cells. C RTPCR of HvAV-3j-encoded iap-1, caspase-like, mcp, glycosyltransferase, helicase, and DNA polymerase in HvAV-3j-inoculated mammalian cells. D RT-PCR of AcMNPVencoded 38k, gp64, polyhedrin, and DNA polymerase in AcMNPV-Egfp-inoculated mammalian cells. M: maker; N: negative control (PCR products from mock infected cellular cDNA); P: positive control (PCR products from vrial genomic DNA).
In the AcMNPV-Egfp infected insect cells, the Dpol (orf65), 38 K (orf98), gp64 (orf128), and polyhedrin (polh, orf8) transcripts were detected only in infected Sf9 and SeFB cells (Fig. 6B). 38k, gp64, and Dpol transcription started at 12 hpi, whereas polyhedrin transcription was detected from 24 hpi (Fig. 6B). No viral transcripts were detected in HvAV-3j- or AcMNPV-Egfp-infected HEK293, 7402, HePG2, TM3, PK15 and ST cells, in line with the above described results.
-
Western blotting assays with a specific polyclonal antibody showed that MCP was expressed at 72 and 96 hpi in HvAV-3j-infected Sf9, HzAM1, SeFB, and HaFB cells (Fig. 7A, 7B), but no target bands were detected in HvAV-3j-infected HEK293, 7402, HePG2, PK15, ST, and TM3 cells (Fig. 7C, 7D). These results indicated that the HvAV-3j proteins were synthesized in Sf9, HzAM1, SeFB, and HaFB cells but not in the tested mammalian cells.
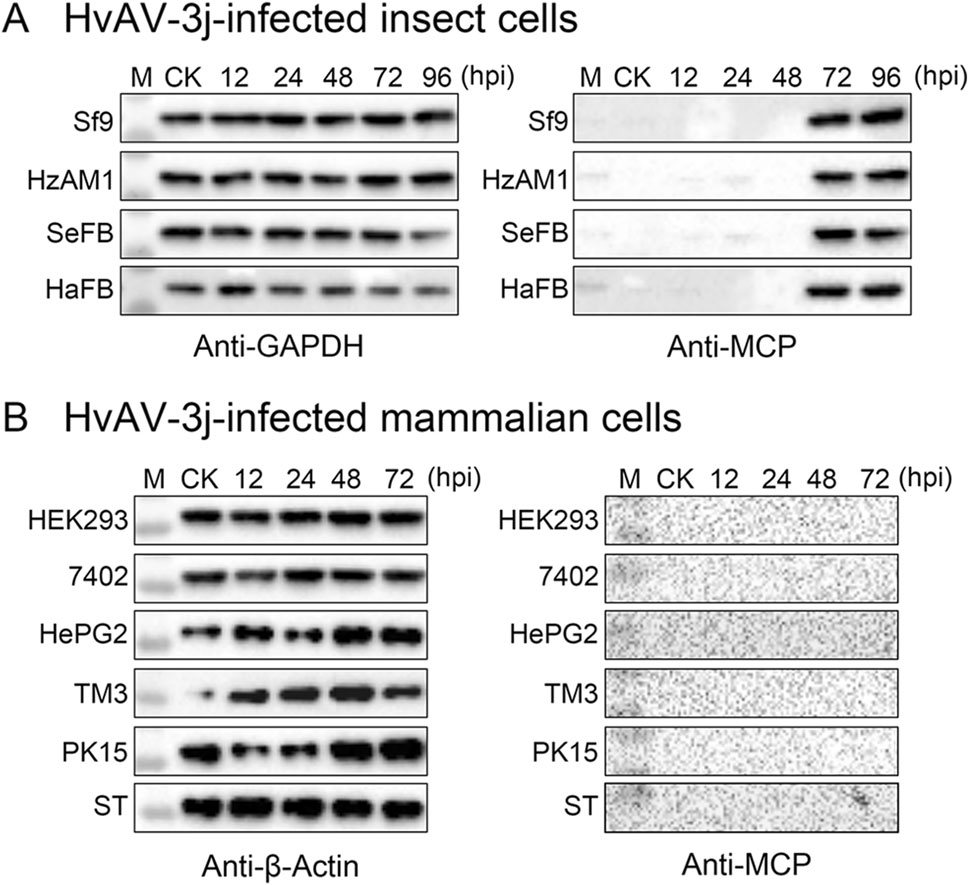
Figure 7. MCP expression detection in HvAV-3j infected cells. A Western blot of HvAV-3j-infected Sf9, HzAM1, SeFB, and HaFB cells with a prepared polyclonal antibody against MCP. A prepared polyclonal antibody against GAPDH was used to detect reference protein expression. B Western blot of HvAV-3j-infected HEK293, 7402, HePG2, TM3, PK15, and ST cells with a prepared polyclonal antibody against MCP. A commercially obtained monoclonal antibody against b-actin was used for detection of the reference protein. M: Marker; CK: Mock-infected cells.
Morphological Changes Were Observed in HvAV-3j-inoculated Insect Cells But Not in HvAV-3j-inoculated Mammalian Cells
Mammalian Cell Proliferation Was Not Affected by HvAV-3j Inoculation
Determination of Viral DNA Replication in Different Cell Lines
Viral Genes Were Not Transcribed in Mammalian Cell Lines
The Major Capsid Protein (MCP) of HvAV-3j Was Exclusively Detectable in Infected Insect Cells
-
In this study, we assessed the risk of mammalian cell infection by HvAV-3j, based on cell morphology and viability, viral DNA replication, as well as transcription and protein expression of different viral genes. In order to compare HvAV-3j-induced effects on cell morphology with those elicited by baculoviruses, a green fluorescence protein-expressing AcMNPV construct was generated and used as a control.
The infection of HvAV-3j caused the enlargement and release of insect cells from the flask on the first day postinfection (Fig. 1A). The HvAV-3j-infected Sf9 and SeFB cells eventually underwent lysis forming vesicles, but this destructive phenomenon was not observed in HzAM1 and HaFB cells, even at 7 dpi (data not shown). Similar events were previously observed with other ascoviruses, as exemplified by the effects of HvAV-3h on SeE1 and Sf9 cells (Huang et al. 2012a), of SfAV-1a on Sf21 cells (Bideshi et al. 2005), and of HvAV-3e on Sf9, Hz, and FB33 cells (Asgari 2006). These results indicated that infection by ascoviruses may lead to different changes in cell morphology in Spodoptera and Helicoverpa. It was reported that the HvAV-3h-infected H. armigera undergo an additional 20 days of larval life, compared to mockinfected larvae, corresponding to a 50% longer larval time than HvAV-3h-infected S. exigua and S. litura larvae (Li et al. 2013). Although it is not known which factors determine the time required for ascovirus-induced larval death, the speed of vesicle formation may play a critical role in this event.
The infection of HvAV-3j aμmost suppressed the proliferation of Sf9, HzAM1, SeFB, and HaFB cells, and the most significant decrease in proliferation took place between 6 and 12 hpi (Fig. 3). Acute pathogenicity of ascovirus was also reported in HvAV-3h-infected Sf9 and SeE1 cells (Huang et al. 2012a). Microscopic observation (Fig. 2) and cell proliferation assays (Fig. 4) indicated that HvAV-3j and AcMNPV did not exert cytotoxic or growthinhibiting effects in human, pig, and mouse cells. Our results were similar to those previously obtained with baculoviruses, showing lack of infectivity toward mammalian cells, which was the precondition for their development into gene therapy vectors (Tjia et al. 1983; Volkman and Goldsmith 1983; Groener et al. 1984; Carbonell et al. 1985).
AcMNPV has a relatively broad host range as compared to other baculoviruses, and is infectious to several lepidopteran larvae. In this study, the constructed AcMNPVEgfp was infectious to Sf9 and SeFB cells, but not to HzAM1 and HaFB cells, which was consistent with a previous study by Thiem (1997). qPCR quantification of viral DNA showed that AcMNPV-Egfp was able to replicate its genomic DNA hundreds of times after 24 hpi, and reached approximately ten thousand genome copies at 96 hpi (Fig. 5B). Studies examining baculoviral DNA replication curves for AcMNPV or HearNPV reported a similar tendency (Wu et al. 2006; Tao et al. 2013; Zhu et al. 2013; Yu et al. 2015). HvAV-3j showed a more efficient DNA replication than AcMNPV-Egfp, producing thousands of DNA copies in 12 h, and millions of copies in 72 h (Fig. 5A). The higher replicative ability of ascoviruses is most likely related to their reproductive capacity, resulting in intracellular vesicle accumulation and causing the clear host larval hemolymph to change into a milky white suspension within the first 3 days post-infection (Govindarajan and Federici 1990). Interestingly, the growth rate of HvAV-3j was similar in all insect cells examined in this study, including cells derived from Heliothis (HzAM1 and HaFB) or Spodoptera (Sf9 and SeFB). This suggests that the observed heterogeneous cytopathological effects were not due to a different viral permissiveness of the tested cell lines.
The analysis of transcription and expression of different viral protein was in total accordance with the results obtained from the analysis of viral DNA replication. HvAV-3j was not infectious to mammalian cells, as no viral protein was expressed in HvAV-3j- or AcMNPVinoculated HEK293, 7402, HePG2, PK15, ST, and TM3 (Figs. 6, 7).
We demonstrated that ascovirus could not infect mammalian cells in vitro, and that the infection by HvAV-3j caused distinct cytopathological alterations in Spodoptera and Helicoverpa cells. Moreover, HvAV-3j infection resulted in early pathogenicity to host insect cells, as assessed by rapid DNA replication and significant decline in cell viability from 6 to 12 hpi. Finally, ascovirus exhibited a more efficient DNA replication as compared to baculovirus. The latter finding evokes the possibility of using ascoviruses as protein expression vectors. However, additional experiments are needed to in-depth characterize the mechanistic basis for the efficient DNA replication of ascovirus. In conclusion, our results indicated that ascoviruses are safe to non-target vertebrate cells, supporting their potential use for the development of biocontrol agents and gene therapy vectors.
-
The authors thank Dr. Zhi-Hong Hu (Wuhan Institute of Virology, Chinese Academy of Sciences) for generously providing HzAM1, Sf9 cells and E. coli strain of DH10Bac; Dr. QiLian Qin (Institute of Zoology, Chinese Academy of Sciences) for generously providing SeFB cells; Dr. Jian-Hong Li (College of Plant Science & Technology, Huazhong Agricultural University) for kindly providing HaFB cells; Dr. Si Qin (College of Food Science and Technology, Hunan Agriculture University) kindly giving HePG2 cells; Dr. Yun Liu (Zunyi Medical University) generously providing 7402 cells; Dr. Zhong Ding (College of Plant Protection, Hunan Agriculture University) generously providing HEK293 cells; Dr. Qing Yang (College of Veterinary Medicine, Hunan Agriculture University) generously providing PK15, ST, and TM3 cells. This work was supported by the National Natural Science Foundation of China (31700141, 31872027) and the grant of Chinese Post-doctoral special funding (2018T110832) and Program to supporting research activities of female researchers in Ministry of education, culture, sports, science and technology-Japan.
-
HY, MN and GHH contributed to the study design. HY, YYOY and NL performed the experiments. HY, MN and GHH contributed reagents. HY and YYOY analyzed the data. HY, YYOY, MN and GHH wrote the manuscript. All authors read and approved the final manuscript.
-
The authors declared no competing financial interests in this paper.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: