-
Dear Editor,
Porcine epidemic diarrhea virus (PEDV) is the etiologic agent of porcine epidemic diarrhea (PED), which is an acute, highly contagious, and devastating enteric viral disease in pigs (Lee 2015). PEDV is a coronavirus that mainly infects and replicates in villous enterocytes of the small intestine in pigs (Li et al. 2016). PEDV can infect pigs in all age groups, with symptoms of varying degrees, such as watery diarrhea, dehydration, vomiting, and growth retardation. Particularly, PEDV is most detrimental to neonatal pigs, and the mortality rates can reach 100% (Stevenson et al. 2013; Lee 2015). PEDV can persist in pigs after infection and become endemic in the farm. PED outbreaks occur periodically in many countries and have resulted in great economic losses to the pig industry worldwide. Effective infection prevention and control of PEDV can only be achieved through the use of vaccines. However, current field epidemics are complex and heterogeneous because of the rapid variation and evolution of PEDV. The genetic diversity between epidemic field strains and vaccine strains could compromise the effectiveness of vaccinations (Li et al. 2012). Moreover, a serious mixed infection with strains from different regions and genogroups is prevalent in China, thereby rendering a difficulty for PEDV prevention (Sun et al. 2015).
An alternative strategy for an effective viral infection control is through genetic modification of a viral receptor gene of the host animals, including gene knockout (KO) or gene replacement with an ortholog from nonpermissive species (Whitworth et al. 2014). The strategy has been validated by a proof-of-concept study in which pigs were conferred resistance against porcine reproductive and respiratory syndrome virus infection by modifying the specific viral receptor gene CD163 (Whitworth et al. 2016). The putative receptor for PEDV infection was considered to be Aminopeptidase N (APN) (Li et al. 2007). Previous reports showed that expression of pig APN in nonsusceptible cell lines rendered susceptibility to PEDV infection (Li et al. 2007). However, the function of APN in PEDV infection remains controversial. Recent studies demonstrated that porcine APN overexpression in nonsusceptible cells failed to support PEDV infection (Shirato et al. 2016; Li et al. 2017), and the depletion or down-regulation of APN expression in PEDV-susceptible cells had no inhibitory effect on PEDV infection (Li et al. 2017; Ji et al. 2018). These cell-based studies demonstrated that APN is not required for PEDV entry to cultured cells. However, discrepancies often exist between the cell level and whole body. The role of APN in PEDV infection in vivo still needs clarification.
To determine whether APN is critical for PEDV infection, we created APN KO pigs by using CRISPR/Cas9- mediated gene editing and somatic cell nuclear transfer (SCNT). Two candidate guide RNAs (gRNAs) were selected in the immediate downstream area of the start codon of APN. One was located in exon 2 and the other was in exon 3 (Fig. 1A). Pig fetal fibroblasts were transfected with a targeting vector based on pX330 (Addgene # 42230) containing a hU6-driven gRNA and a CAG-driven humanized Cas9. Transfected cells were grown in a low density to form single-cell colonies, which were individually cultured and genotyped to identify the positive ones. A total of 72 and 50 well-propagated single-cell colonies were collected from exon 2- and exon 3-targeting groups, respectively. Among these colonies, 60 and 45 presented targeted insertions and/or deletions (indels) with various patterns in the genome. Four biallelic APN KO colonies, namely, # 2–1, # 2–2, # 2–5 that carried out-of-frame indels in exon 2, and # 3–6 that carried out-of-frame indel in exon 3, were used as nuclear donors for SCNT (Supplementary Table S1). A total of 2, 494 embryos were constructed from the selected positive cells and transferred into 11 surrogates. Four surrogates were pregnant to term and delivered 18 live piglets (2 died at day 1 due to weakness at birth) and 5 stillborns (Fig. 1B and Supplementary Table S2). Genotyping of the live cloned piglets showed that all of them were biallelic or homozygous KO with out-of-frame indels in exon 2 of APN (Supplementary Table S3 and Supplementary Fig. S1). Intriguingly, the cloned piglet groups derived from # 2–1 were divided into two genotypes, homozygous 1-bp deletion (FF23, FF30, FF35, FF36) and homozygous 1-bp insertion (FF21, FF31). This result indicated that # 2–1, which was originally considered biallelic KO with a 1-bp deletion (∆1) in one allele and a 1-bp insertion (+1) in the other allele, was definitely a mixed colony composed of two homozygous KO colonies. We next determined the APN protein expression status in the cloned piglets. Western blot with a rabbit anti-APN antibody (ab183358, Abcam) showed no APN expression in the small intestines of two dead piglets (# 1 & # 2 with ∆1/∆7 genotypes), whereas wild-type (WT) control displayed significant APN expression level (Fig. 1C).
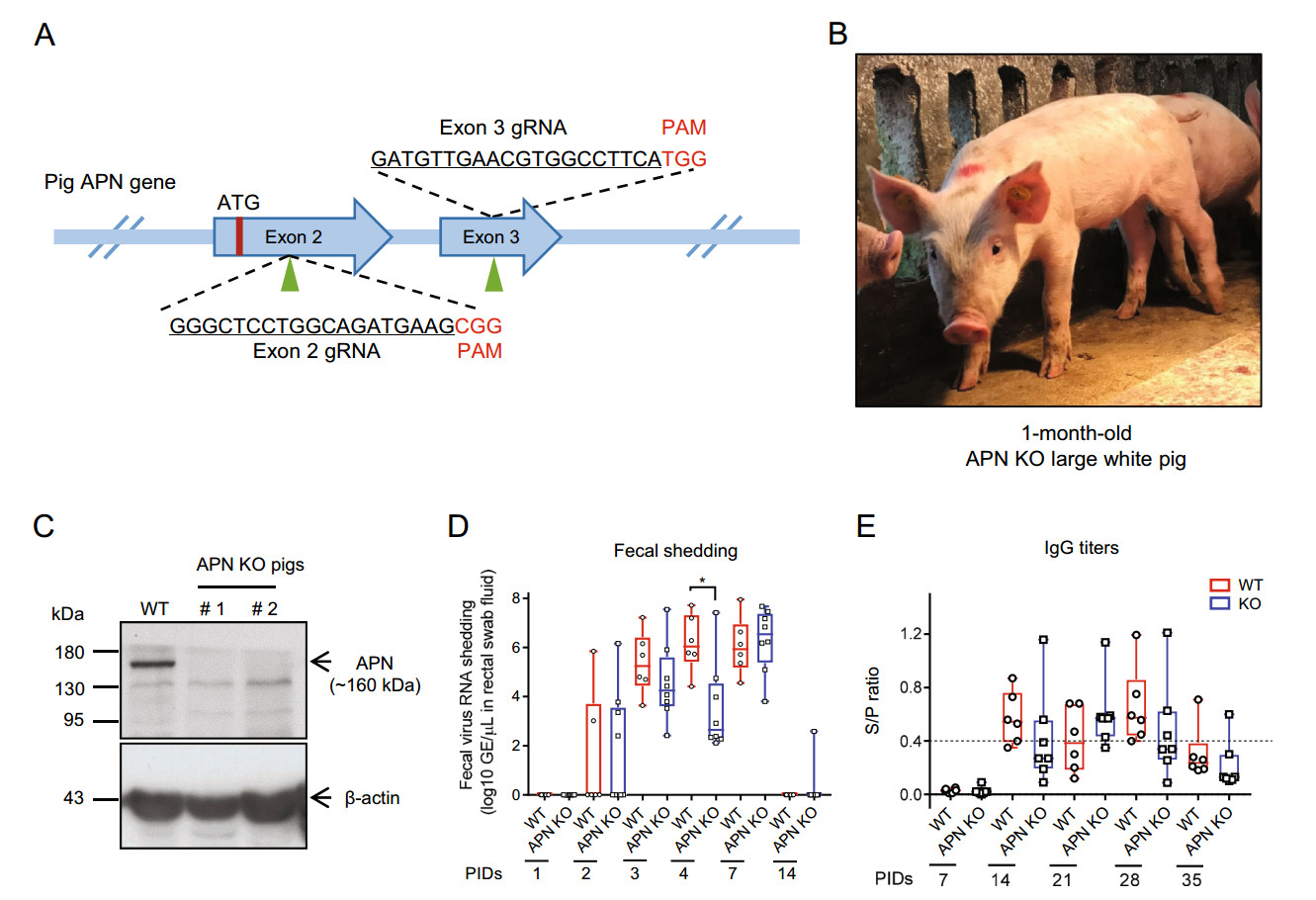
Figure 1. A Pig APN gene targeting strategy. Schematic of the pig APN locus showing the location (indicated by arrowheads) and sequence (underlined) of two gRNAs targeting exons 2 and 3 individually. Red letters denote the protospacer adjacent motif (PAM). B Representative photo of APN KO cloned pig (1 month old). C Western blot showing loss of APN expression in the intestines of KO pigs compared with the significant APN expression in WT pig. β-actin is used as the loading control. D Quantitative PCR detection of PEDV RNA in fecal swabs. Fecal shedding of both groups is first observed at PID 2, present in all the pigs with peak titers more than 7 log10 GE/lL in individual pigs at PIDs 3, 4, and 7, and absent in most pigs at PID 14. No significant difference in the fecal shedding was found between KO and WT group, excluding that at PID 4 (*P < 0.05, unpaired t test). E Antibody response after PEDV challenge in the experimental groups. The two groups did not differ in the antibody titers. An S/P ratio higher or equal to 0.4 (broken line) was considered positive. Data in D and E are expressed with box and whiskers plot around median values.
PEDV challenge was performed according to a previous time-course study using 4-week-old feeder pigs (Niederwerder et al. 2016). A PEDV field isolate (CT, GenBank Accession No. MK539948) from South China was propagated on Vero cells and the 10th generation of PEDV stock (6.0 Log10 TCID50/mL) was used for PEDV challenging in pigs. A total of 9 APN KO and 6 WT male Large White pigs at 30 days old were orally infected with 6 mL PEDV solution. The clinical sign of each pig was observed daily following the challenge. Diarrhea was assessed by fecal consistency level, which was scored as follows: 0 = solid, 1 = pasty, 2 = semiliquid, and 3 = liquid. A score of 2 or 3 was considered diarrheic. Fecal swabs of each challenged pig were collected at post-inoculation days (PIDs) 0, 1, 2, 3, 4, 7, and 14 to detect the viral shedding. The blood was drawn at PIDs 0, 7, 14, 21, 28, and 35 to isolate serum for antibody and viremia detection. Following infection, only a short period of diarrhea (2–3 days) was observed from portions of both WT and KO pigs, and no significant difference in clinical signs was found between the WT and APN KO pigs (Supplementary Table S4). Two APN KO pigs manifested consistent diarrhea and evident body weight loss, and died at PIDs 3 and 7, respectively. The other pigs remained active and maintained good bodily condition after the diarrhea resolved.
To analyze PEDV RNA titers in fecal samples, the fecal material on the swabs was squeezed out to the saline and subjected to viral RNA extraction. Viral RNA was reversetranscribed to cDNA by using PrimeScriptTM RT Master Mix (Takara). Thereafter, the viral copy numbers were assayed by quantitating the PEDV nucleocapsid (N) gene by using THUNDERBIRD® Probe qPCR Mix (TOYOBO), and the primers and probe were as follows: PED-N-Forward, 5'-GAATTCCCAAGGGCGAAAAT-3', PED-N-Reverse, 5'-TTTTCGACAAATTCCGCATCT-3', and PED-N-Probe, FAM-CGTAGCAGCTTGCTTCGGACCCA-BHQ1 (Miller et al. 2016). Results showed that the fecal shedding of PEDV RNA could be detected starting from PID 2, and all the pigs shed virus at PIDs 3, 4, and 7. Although the median value of viral titers in feces of KO group was significantly lower than that of WT group at PID 4, the virus shedding levels were not different at other time points. Most animals were negative for fecal shedding at PID 14 (Fig. 1D). The PEDV infection levels were further evaluated by testing the PEDV-specific antibody (IgG) level in the serum with Swinecheck® PED indirect ELISA (Biovet) according to the manufacturer's protocol. Serum PEDV antibody was negative in all the inoculated pigs at and before PID 7. Positive antibody level was observed in 2/7 KO and 4/6 WT pigs at PID 14. An equally peak antibody titer was observed in individual pigs of both groups at PID 28. Most pigs were negative for antibody response at PID 35 (Fig. 1E). Moreover, no inoculated pigs had detectable PEDV RNA in the serum over the infection period by quantitative PCR.
During the completion of our studies a paper published by Dr. Prather group reported the same modified pigs by zygote injection of CRISPR RNAs (Whitworth et al. 2019). These pigs showed resistance to porcine transmissible gastroenteritis virus (TGEV) infection. However, only one APN null pig was used in the PEDV challenge test and the results indicated that the APN null pig remained infected by PEDV. Given that APN plays a putative role as a receptor in PEDV infection, a detailed study should be undertaken to compare the difference in infection levels between WT and KO pigs to assess any resistance to PEDV conferred by APN ablation. Our results demonstrated that APN KO and WT pigs had similar clinical signs, fecal shedding and serum antibody levels, thereby excluding its role in PEDV infection. Nonetheless, APN has been identified as a functional receptor for multiple other coronaviruses (Li et al. 2018), thereby suggesting the importance of APN KO pigs in determining the receptor role of APN for these coronaviruses and in further establishing coronavirus-resistant pigs.
HTML
-
We acknowledge the staffs of cloning lab in Wens Corp for assistance in pig cloning, and Wei Li of Wens Academy for assistance in PEDV culture and challenge. The work was supported by research grant from Science and Technology Planning Project of Guangdong Province for establishing PEDVresistant pigs (2017B020201009).
-
The authors declare that they no conflict of interest.
-
All procedures for creating the pigs and viral challenge experiments were approved by the IACUC at South China Agricultural University.















 DownLoad:
DownLoad: