HTML
-
The family Baculoviridae encompasses a diverse group of insect-specific viruses with circular double-stranded DNA genomes packaged within enveloped, rod-shaped nucleocapsids (Harrison et al. 2018). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been studied most extensively and is regarded as the model baculovirus (Blissard and Theilmann 2018). A typical characteristic of AcMNPV and other relatively well-characterized lepidopteran NPVs is a biphasic life cycle with the production of two morphologically distinct but genetically identical progeny virions, budded virions (BVs) and occlusion-derived virions (ODVs) (Jehle et al. 2006; Rohrmann 2013). The major differences between BVs and ODVs are the source and composition of their envelopes, which may parallel their different functional roles in the baculovirus life cycle (Braunagel and Summers 1994; Slack and Arif 2007; van Oers and Vlak 2007). BVs are responsible for spreading the infection within a larva or among cells in culture, and they acquire envelopes from viral protein-modified areas of the plasma membrane via a strategy similar to that of other viruses that bud from the cell surface (Blissard and Wenz 1992; Hong et al. 1997; Braunagel and Summers 2007; Slack and Arif 2007). ODVs can initiate primary infections in the midgut epithelium of infected insects and are thus required for interhost transmission (Hodgson et al. 2007; Slack and Arif 2007). ODVs obtain their envelopes from virus-induced intranuclear microvesicles within the nucleoplasm (Braunagel and Summers 2007; Slack and Arif 2007; Rohrmann 2013; Blissard and Theilmann 2018). The molecular mechanism of ODV morphogenesis remains unclear.
The assembly of ODVs requires a complex integration of events that include the formation of the precursors of ODV envelopes, referred to as intranuclear microvesicles, followed by the envelopment of nucleocapsids at these intranuclear microvesicles (Blissard and Theilmann 2018). The mechanism that underlies the morphogenesis of intranuclear microvesicles remains to be clarified. Although there has been some controversy regarding the source of intranuclear microvesicles, accumulating studies have provided strong evidence to support the hypothesis that the formation of intranuclear microvesicles is the result of budding of the ODV envelope protein-decorated inner nuclear membrane (INM) into the nucleoplasm (Williams and Faulkner 1997; Braunagel and Summers 2007; Slack and Arif 2007). Therefore, the production of intranuclear microvesicles could be divided into two steps: (1) trafficking of ODV membrane proteins to the nuclear membrane and (2) budding of the nuclear membrane into the nucleoplasm to form intranuclear microvesicles (Braunagel and Summers 2007; Hellberg et al. 2016). The transport of ODV INM-directed integral proteins from the endoplasmic reticulum to the INM has been well studied, and two viral proteins, FP25-K and BV/ODV-E26, and the cellular protein importin-α-16 participate in this pathway (Braunagel et al. 2004, 2009). However, the mechanism involved in the budding of the nuclear membrane has just started to be investigated in detail. Three viral genes, ac75, ac76, and ac93, have recently been reported to be required for intranuclear microvesicle formation (Hu et al. 2010; Yuan et al. 2011; Shi et al. 2018). The deletion of each of these genes results in a deficiency of intranuclear microvesicles. Following the formation of intranuclear microvesicles, nucleocapsids retained within the nucleus associate with these intranuclear microvesicles and are consequently enveloped to form ODVs (Williams and Faulkner 1997; Braunagel and Summers 2007). Four viral genes have been identified as required for this process, including ac11, ac94, ac109, and ac142. The deletion of any of the above genes does not affect nucleocapsid assembly and intranuclear microvesicle formation but rather abrogates the envelopment of nucleocapsids at intranuclear microvesicles and results in the absence of ODVs (McCarthy et al. 2008; Chen et al. 2012; Lehiy et al. 2013; Tao et al. 2015).
p48 is a core gene that has been identified in all sequenced baculovirus genomes (Garavaglia et al. 2012). p48 was originally identified as an open reading frame (ORF) within the Orgyia pseudotsugata MNPV (OpMNPV) Hind Ⅲ G fragment and predicted to encode a gene product of 48 kDa (Russell and Rohrmann 1990). AcMNPV p48 (ac103) is a late gene that is predicted to encode a protein of 387 amino acids with a putative molecular mass of 45 kDa (Ayres et al. 1994; Chen et al. 2013). Analysis of the expression of P48 suggested that proteolytic cleavages may occur at the N terminus of P48 (Yuan et al. 2010). The deletion of p48 does not affect viral DNA replication, nucleocapsid assembly, or the transport of nucleocapsids from the virogenic stroma (VS) to the nuclear ring zone. However, this deletion precludes the nuclear egress of nucleocapsids, formation of ODVs, and the subsequent formation of BVs and embedding of ODVs into polyhedra (Yuan et al. 2008). Two recent studies showed that the host membrane fusion and scission machineries, including the N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) machinery, which triggers membrane fusion, and the endosomal sorting complex required for transport-Ⅲ (ESCRT-Ⅲ) complex, which is responsible for membrane scission, are involved in the nuclear egress of nucleocapsids, and P48 associates with the NSF protein of the SNARE machinery and the Vps24 subunit of the ESCRT-Ⅲ complex (Guo et al. 2017; Yue et al. 2018). These results suggested that P48 may affect the nuclear egress of nucleocapsids by associating with the SNARE and ESCRT machineries. However, the exact role played by p48 in ODV morphogenesis remains unknown.
In the present study, we present evidence that P48 is required for the efficient formation of intranuclear microvesicles. P48 is associated with both the nucleocapsid and envelope fractions of BVs and ODVs. In virus-infected cells, P48 is predominantly localized to the nucleocapsids in the VS and the nucleocapsids enveloped in ODVs, and this protein is also detected in the plasma membrane, nuclear envelope, intranuclear microvesicles, and ODV envelope. P48 associates with Ac93, which is also involved in intranuclear microvesicle formation, independent of other viral proteins.
-
Spodoptera frugiperda IPLB-Sf21-AE clonal isolate 9 (Sf9) cells (Vaughn et al. 1977) were cultured at 27 ℃ in Grace's insect medium (Thermo Fisher Scientific, Waltham, US) supplemented with 10% fetal bovine serum, penicillin (100 μg/mL), and streptomycin (30 μg/mL). Larvae of Trichoplusia ni (T. ni) were reared on an artificial diet at 28 ℃ (Li et al. 2002). The wild-type AcMNPV vAcWT was constructed by the insertion of the polyhedrin (polh) gene and green fluorescent protein (GFP) encoding gene into the polh locus of the bacmid bMON14272 (Cai et al. 2012). The BV titer was determined by using a 50% tissue culture infective dose (TCID50) endpoint dilution assay (EPDA) on Sf9 cells (O'Reilly et al. 1992).
The polyclonal antibody against AcMNPV VP39 was generated by Li et al. (2007), and the polyclonal anti-AcMNPV ODV-E25 antibody was a gift from Prof. Zhihong Hu (Wuhan Institute of Virology, CAS, China) (Wang et al. 2010). The mouse monoclonal anti-FLAG antibody, mouse monoclonal anti-Myc antibody, and 50% slurry of protein A/G agarose beads conjugated to an anti-Myc antibody or anti-FLAG antibody were purchased from Abmart (Shanghai, China). The mouse monoclonal anti-GP64 AcV5 antibody was purchased from (Thermo Fisher Scientific). Goat anti-mouse IgG-conjugated 10-nm gold particles were purchased from Sigma-Aldrich (St. Louis, US). The horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody and donkey anti-rabbit antibody were purchased from Thermo Fisher Scientific and GE Healthcare (Chicago, US), respectively.
-
To generate a FLAG-tagged P48 repair bacmid, a donor plasmid containing a 3×FLAG-encoding sequence at the 3' end of p48 was constructed. The p48 gene fragment containing its native promoter sequence was PCR-amplified from bMON14272 with primers EcoRI-PP48-BamHI-F and P48-XbaI-R (the PCR primers used in this study are listed in Supplementary Table S1). The PCR product was digested with EcoR I and Xba I and cloned into pUC18-3FSV40, which contains a 3×FLAG-encoding sequence and a simian virus 40 (SV40) poly(A) signal (Shi et al. 2018), to generate pUC18-P48:FLAG-SV40. The fragment P48:FLAG-SV40 was digested from plasmid pUC18-P48:FLAG-SV40 and subcloned into pFB1-PH-GFP (Wu et al. 2006) to generate the donor plasmid pFB1-P48:FLAG-PH-GFP. The donor plasmid was then transformed into DH10B electrocompetent cells containing the pMON7124 helper plasmid and a p48-knockout bacmid, bP48KO, to generate the recombinant virus vP48:FLAG as previously described (Wu et al. 2006).
To determine whether P48 associates with the proteins required for intranuclear microvesicle formation in the absence of viral infection, plasmids transiently expressing Ac75, Ac76, Ac93, and P48 were constructed for immunoprecipitation assays. Briefly, the p48 ORF, with a Kozak consensus sequence for the proper initiation of translation (Kozak 1987, 1991) at its 5' end and a FLAGcoding sequence at its 3' end, was amplified from bMON14272 using primers KP48-BamHI-F and P48:FXbaI-R and then cloned into the transient-expression vector pIB/V5-His (Thermo Fisher Scientific) to generate pIBP48:FLAG. The ORFs of ac75, ac76, ac93, and p48 with Kozak consensus sequences at their 5' ends and a Myccoding sequence at their 3' ends were amplified from bMON14272 with the primer pairs KAc75-BamHI-F/ Ac75:M-XbaI-R, KAc76-BamHI-F/Ac76:M-XbaI-R, KAc93-BamHI-F/Ac93:M-XbaI-R, and KP48-BamHI-F/ P48:M-XbaI-R. The resulting PCR products were digested with BamHI and XbaI and cloned into pIB/V5-His to generate pIB-Ac75:Myc, pIB-Ac76:Myc, pIB-Ac93:Myc, and pIB-P48:Myc, respectively.
Recombinant wild-type viruses expressing Myc-tagged Ac75, Ac76, Ac93, and P48 as well as FLAG-tagged P48 were generated to determine the association between P48 and Ac75, Ac76, Ac93, or itself during viral infection. The Myc-tagged ac75, ac76, ac93, and p48 ORFs with their native promoter sequences were amplified from bMON14272 with primer pairs PAc75-EcoRI-F/Ac75:MBamHI-R, PAc76-EcoRI-F/Ac76:M-BamHI-R, PAc93-EcoRI-F/Ac93:M-BamHI-R, and PP48-EcoRI-F/P48:M-BamHI-R. The PCR products were cloned into plasmid pUC18-SV40 (Cai et al. 2012) to generate pUC18-Ac75:Myc-SV40, pUC18-Ac76:Myc-SV40, pUC18-Ac93:Myc-SV40, and pUC18-P48:Myc-SV40. These plasmids were digested with EcoR I and Pst I, and the resulting fragments containing the above-mentioned ORFs as well as their native promoters and an SV40 poly(A) signal were subcloned into pFB1-PH-GFP to generate donor plasmids pFB1-Ac75:Myc-PH-GFP, pFB1-Ac76:Myc-PH-GFP, pFB1-Ac93:Myc-PH-GFP, and pFB1-P48:Myc-PH-GFP. The above donor plasmids and pFB1-P48:FLAG-PH-GFP were transformed into electrocompetent DH10Bac cells (Thermo Fisher Scientific) harboring the pMON7124 helper plasmid and bMON14271 to generate the recombinant viruses vAcWT-Ac75:Myc, vAcWT-Ac76:Myc, vAcWT-Ac93:Myc, vAcWT-P48:Myc, and vAcWT-P48:FLAG.
All of the constructs were verified by PCR analysis and DNA sequencing. Bacmid DNA and transient-expression plasmids were isolated with the Qiagen large-construct kit (Qiagen, Hilden, Germany) and Endo-free Plasmid Mini Kit Ⅱ (Omega Bio-tek, Norcross, US), respectively, and were quantified by determining the optical density.
-
Sf9 cells (1 × 106 cells/35-mm-diameter dish) were transfected with 2.0 μg of bacmid vAcWT or vP48KO by using Cellfectin Ⅱ reagent (Thermo Fisher Scientific). The cells were dislodged at 72 h posttransfection (p.t.), pelleted at 500 ×g for 10 min, and prepared for TEM as previously described (Li et al. 2005a). The samples were observed with a JEM-100CX/II transmission electron microscope at an accelerating voltage of 100 kV.
-
BVs and ODVs were purified from vP48:FLAG-infected fourth-instar T. ni larvae and fractionated into envelope and nucleocapsid fractions as previously described (Braunagel and Summers 1994; Wu et al. 2008). Western blotting with one of the following primary antibodies was performed according to the manufacturer's instructions: (1) mouse monoclonal anti-FLAG antibody (1:2000), (2) rabbit polyclonal anti-AcMNPV VP39 antiserum (1:1000), (3) rabbit polyclonal anti-AcMNPV ODV-E25 antiserum (1:2000), or (4) mouse monoclonal anti-GP64 AcV5 antibody (1:3000). An HRP-conjugated donkey anti-rabbit antibody (1:10, 000) or goat anti-mouse antibody (1:7000) was used as the secondary antibody.
-
Sf9 cells (1.0 × 106) were infected with vP48:FLAG at an MOI of 10 TCID50/cell and harvested at 60 h postinfection (p.i.). The cells were prepared for IEM using LR White resin (Ted Pella, Inc., Redding, US) as previously described (Wei et al. 2014). The ultrathin sections were immunogold labeled as previously described (Wu et al. 2008) with a mouse monoclonal anti-FLAG antibody (1:80), followed by incubation with goat anti-mouse IgG-conjugated 10-nm gold particles (1:50) as the secondary antibody. The samples were visualized with a JEM-100CX/11 transmission electron microscope at an accelerating voltage of 100 kV.
-
Coimmunoprecipitation assays were performed as previously described (Guan et al. 2016), with some modifications. To detect the association between P48 and the proteins that participate in intranuclear microvesicle formation, Sf9 cells (2.0 × 107) were cotransfected with 50 μg of pIB-P48:FLAG combined with 30 μg of plasmids pIB-Ac75:Myc, pIB-Ac76:Myc, pIB-Ac93:Myc, or pIB-P48:Myc. Sf9 cells cotransfected with 50 μg of pIB-P48:FLAG and 30 μg of pIB-V5/His were used as a negative control. To achieve a higher transfection efficiency, a second cotransfection was performed at 18 h after the first cotransfection. At 36 h after the second cotransfection, the cells were harvested and lysed in cell lysis buffer for Western blotting and immunoprecipitation (20 mmol/L Tris [pH 7.5], 150 mmol/L NaCl, and 1% Triton X-100; Beyotime Biotechnology, Shanghai, China) supplemented with 2 μg/mL cOmplete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). After incubation on a vertical rotating mixer for 5 h, the cell lysates were centrifuged at 14, 000 ×g for 10 min at 4 ℃, and the supernatants were transferred to a fresh Eppendorf tube and mixed with 80 μL of a 50% slurry of protein A/G agarose beads conjugated to an anti-Myc antibody. After incubation at 4 ℃ for 14 h while rotating, the beads were collected by centrifugation at 2000 ×g for 3 min at 4 ℃, washed three times with 1 mL of the lysis buffer and boiled in 90 μL of 1× loading buffer for 10 min. The dissolved immunoprecipitates and the input cell lysates were analyzed by Western blotting with a mouse monoclonal anti-FLAG (1:2000) or anti-Myc (1:2000) antibody.
To determine the association between P48 and Ac75, Ac76, Ac93 or P48 itself during viral infection, Sf9 cells (1.0 × 107) were coinfected with vAcWT-P48:FLAG combined with vAcWT-Ac75:Myc, vAcWT-Ac76:Myc, vAcWT-Ac93:Myc, or vAcWT-P48:Myc (each at an MOI of 5 TCID50/cell). At 36 h p.i., the cells were collected and lysed for coimmunoprecipitation assay as described above. Sf9 cells coinfected with vAcWT-P48:FLAG and vAcWT were used as a negative control. Western blotting was performed with a mouse monoclonal anti-FLAG or antiMyc antibody.
Cells, Insects, Viruses, and Antibodies
Construction of Viruses and Plasmids
Transmission Electron Microscopy (TEM)
BV and ODV Purification
Immunoelectron Microscopy (IEM)
Coimmunoprecipitation
-
Our previous study showed that the deletion of p48 blocked ODV formation but did not affect nucleocapsid assembly (Yuan et al. 2008). There may be two possibilities: (1) p48 is involved in the formation of intranuclear microvesicles, which are precursors of ODV envelopes, and (2) the deletion of p48 interferes with the envelopment of nucleocapsids at intranuclear microvesicles. To determine the exact role of p48 in ODV formation, TEM was performed on cells transfected with the recombinant wild-type bacmid vAcWT or the p48 knockout bacmid vP48KO (vAcP48-KO-PH-GFP) (Yuan et al. 2008) at 72 h p.t. As expected, numerous virus-induced intranuclear microvesicles were observed in vAcWT-transfected cells (Fig. 1A-a). However, the number of intranuclear microvesicles observed in vP48KO-transfected cells was significantly reduced. No intranuclear microvesicles were observed in more than half of vP48KO-transfected cells (data not shown), and only a few intranuclear microvesicles were observed in the remaining cells (Fig. 1A-b). To generate a quantitative estimate of intranuclear microvesicle production, 20 transfected cells in thin sections of each sample were randomly chosen, and the total number of intranuclear microvesicles was counted. As shown in Fig. 1B, the number of intranuclear microvesicles derived from vP48KO-transfected cells (15 ± 1 per 20 cells) was reduced by over 99.7% (P = 0.0003) compared with that from vAcWT-transfected cells (4820 ± 695 per 20 cells). This result indicated that the deletion of p48 severely decreased intranuclear microvesicle formation.

Figure 1. TEM of Sf9 cells transfected with vAcWT or vP48KO. A Representative images showing the intranuclear microvesicles (white arrows) in Sf9 cells transfected with vAcWT (a) or vP48KO (b) at 72 h p.t. Nu, nucleus; Cy, cytoplasm; NE, nuclear envelope. Scale bar, 500 nm. B Quantitative analyses of intranuclear microvesicle production. Twenty cells were randomly selected from thin sections of vAcWT or vP48KO-transfected cells, and the total number of intranuclear microvesicles was counted. The values represent the average from three independent assays. The error bars indicate the standard deviations. The P value was calculated using the Student's t test. ***, P < 0.001.
-
The proteins that have been identified as involved in intranuclear microvesicle formation thus far, including Ac75, Ac76, and Ac93, are all viral structural proteins (Yuan et al. 2011; Wei et al. 2014; Shi et al. 2018). To further investigate the role of P48 in intranuclear microvesicle formation, it is necessary to determine whether P48 is associated with the virion structure. Thus, a FLAG-tagged P48 repair bacmid, vP48:FLAG, was generated by inserting the gene encoding C-terminally FLAG-tagged P48, and the genes for GFP and polyhedrin into the polh locus of a p48-knockout bacmid, bP48KO (vAcP48-KO) (Yuan et al. 2008), so that P48 could be monitored with a commercially available antibody (Fig. 2A). The propagation of vP48:FLAG and vAcWT in Sf9 cells was compared to determine the effect of C-terminally FLAG-tagged P48 on viral replication. The supernatants of vP48:FLAG- or vAcWT-transfected Sf9 cells were collected at 96 h p.t., and the BV titers were determined by a TCID50 EPDA. The titer of vP48:FLAG was similar to that of vAcWT (Fig. 2B), indicating that vP48:FLAG and vAcWT had a similar ability to produce infectious BVs. Additionally, TEM analysis was performed to determine whether FLAG-tagged P48 has any effect on viral morphogenesis. As expected, both vP48:FLAG- and vAcWT-transfected cells showed typical cytological changes and viral morphogenesis (data not shown), indicating that FLAG-tagged P48 was able to rescue the defective morphogenesis of vP48KO. The above results demonstrated that C-terminally FLAG-tagged P48 behaved similarly to the original P48 and is appropriate for subsequent analyses.
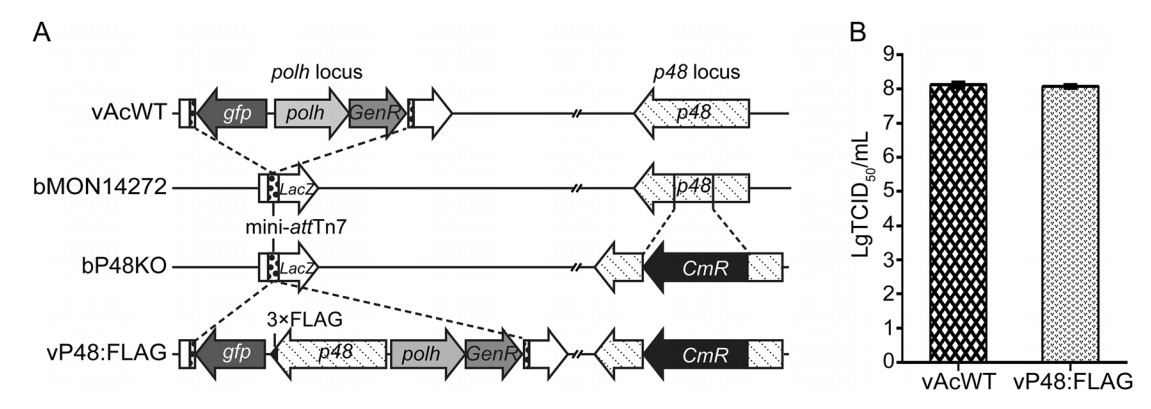
Figure 2. Construction of the FLAG-tagged P48 repaired virus and analysis of its fitness. A Schematic diagram of the polh and p48 insertion loci of vAcWT, bP48KO, and vP48:FLAG. The FLAG-tagged P48 repaired virus (vP48:FLAG) was constructed by inserting the p48 gene tagged with a 3 × FLAG epitope sequence (black triangle) at its 3' end, under the control of its native promoter, and the polh and gfp genes into the polh locus of bP48KO. The vAcWT bacmid was generated by the insertion of the polh and gfp genes into the polh locus of bMON14272. GenR, gentamicin resistance; CmR, chloramphenicol resistance. B Analysis of infectious BV production of vP48:FLAG and vAcWT. Sf9 cells were transfected with the bacmid vP48:FLAG or vAcWT. The supernatants were harvested at 96 h p.i., and BV titers were determined using TCID50 EPDA. Each value represents the average titer derived from three independent transfections. The error bars indicate the standard deviations.
Fourth-instar T. ni larvae were infected with vP48:FLAG polyhedra. The BVs and ODVs were purified from the infected larvae, fractionated into nucleocapsid and envelope fractions, and analyzed by Western blotting. As shown in Fig. 3, FLAG-tagged P48 was detected in both the nucleocapsid and envelope fractions of BVs and ODVs. As a control to confirm that there was no cross-contamination between the fractions, the major nucleocapsid protein VP39, the BV/ODV envelope protein ODV-E25, and the BV envelope-specific protein GP64 were also analyzed by Western blotting, and all of these three proteins were detected only in the expected fractions. These results indicated that P48 is associated with BVs and ODVs and sublocalized to the nucleocapsid and envelope fractions of both virions.
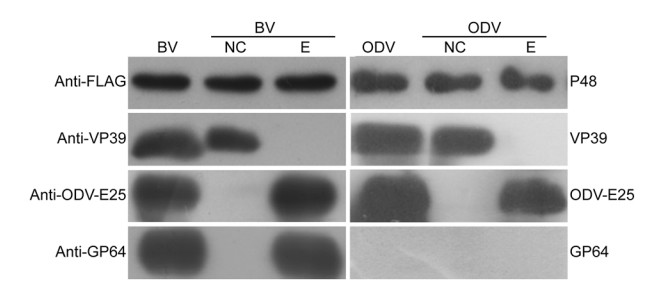
Figure 3. Western blot analysis of P48 in purified and fractionated virions. BVs and ODVs were purified from vP48:FLAG-infected larvae and fractionated into envelope and nucleocapsid fractions. Immunoblotting was performed with an anti-FLAG antibody to detect FLAG-tagged P48, an anti-VP39 antibody to detect the major capsid protein VP39, an anti-ODV-E25 antibody to detect the BV/ODV envelope-associated protein ODV-E25, and an anti-GP64 antibody to detect the BV envelope-specific protein GP64. NC, nucleocapsid fraction; E, envelope fraction.
-
To further determine the fine structure localization of P48 in virus-infected cells, vP48:FLAG-infected cells were collected at 60 h p.t. and prepared for IEM with a mouse monoclonal anti-FLAG antibody as the primary antibody. Immunogold particles were predominantly detected in both naked nucleocapsids in the VS (Fig. 4A) and the nucleocapsids enveloped in ODVs (Fig. 4B, black arrow). Additionally, limited but discernible immunogold particles were also observed to be associated with the envelope of ODVs (Fig. 4B, white arrow), intranuclear microvesicles (Fig. 4C), the nuclear envelope (Fig. 4D), and the plasma membrane (Fig. 4E). As a negative control, there were nearly no immunogold particles present in vAcWT-infected cells (data not shown). The localization pattern of P48 was consistent with the results obtained from the viral fractionation study, which showed that P48 is associated with both the nucleocapsid and envelope fractions of BVs and ODVs.
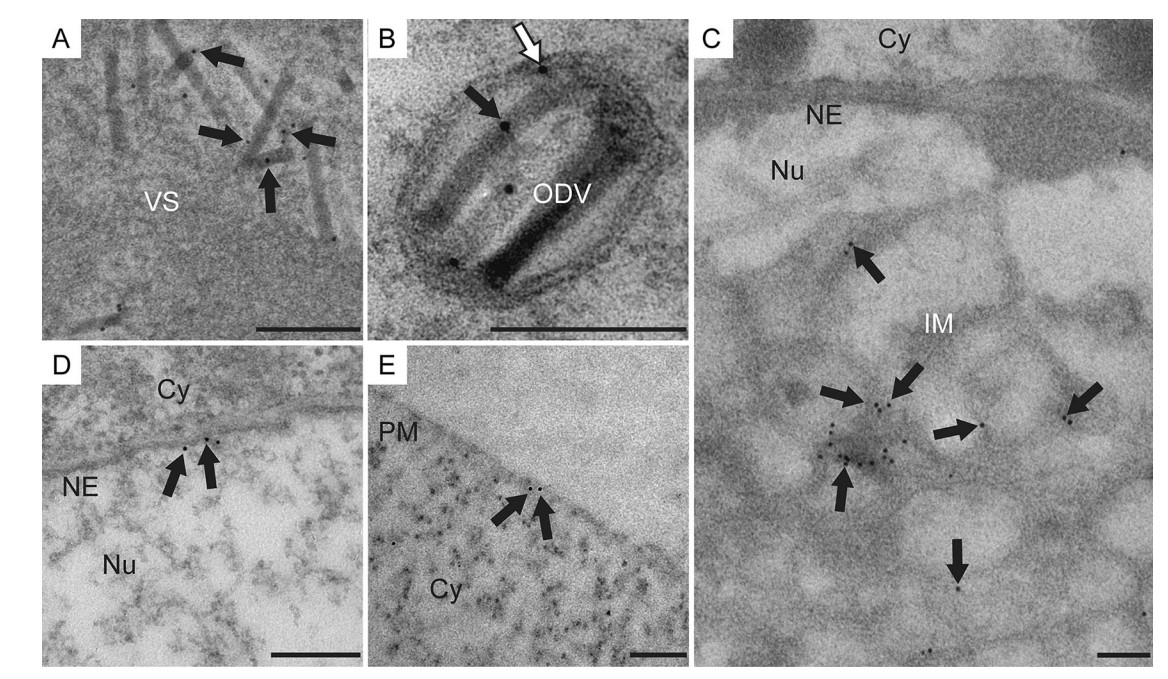
Figure 4. Immunogold localization of P48 in vP48:FLAG-infected cells. Sf9 cells infected with vP48:FLAG at an MOI of 10 TCID50/cell were harvested at 60 h p.i. and processed for immunogold labeling with a mouse monoclonal anti-FLAG antibody. The arrows indicate the immunogold labeling of P48 on the nucleocapsids in the VS (A), nucleocapsid (black arrow) and envelope (white arrow) of ODVs (B), intranuclear microvesicles (C), nuclear envelope (D), and the plasma membrane (E). Cy, cytoplasm; Nu, nucleus; PM, plasma membrane; NE, nuclear envelope. Scale bars, 200 nm.
-
As individual knockouts of ac75, ac76, or ac93 result in the blockade of intranuclear microvesicle formation (Hu et al. 2010; Yuan et al. 2011; Shi et al. 2018), which is similar to the defects caused by the knockout of p48, it is necessary to determine whether P48 associates with these proteins or with P48 itself. Thus, coimmunoprecipitation assays were performed on the above proteins. Sf9 cells were cotransfected with the transient-expression plasmids pIB-P48:FLAG and pIB-Ac75:Myc, pIB-Ac76:Myc, pIB-Ac93:Myc, or pIB-P48:Myc. To obtain a higher yield of the target proteins, a second cotransfection was performed at 18 h after the first cotransfection. At 36 h after the second cotransfection, the cells were harvested and subjected to immunoprecipitation with protein A/G agarose beads conjugated to an anti-Myc antibody. Cells cotransfected with pIB-P48:FLAG and pIB-V5/His were used as a negative control. Western blotting confirmed the expression of each protein (Fig. 5A, Input lanes) and the immunoprecipitation ofAc75:Myc, Ac76:Myc, Ac93:Myc, and P48:Myc by beads conjugated to an anti-Myc antibody (Fig. 5A, IP: anti-Myc lanes). P48 was coimmunoprecipitated with Ac93:Myc but failed to be immunoprecipitated with Ac75:Myc, Ac76:Myc or P48:Myc (Fig. 5A, IP: anti-Myc lanes), indicating that P48 associates with Ac93 in the absence of viral infection.
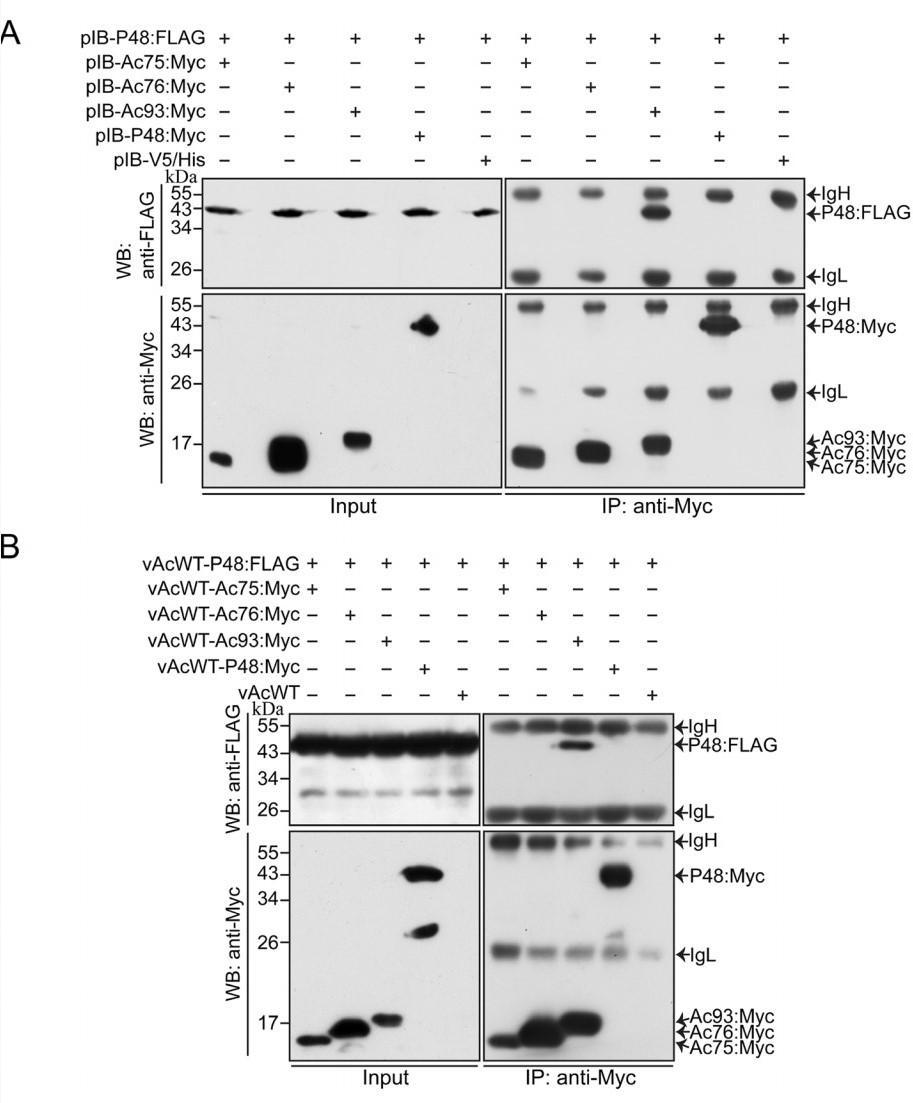
Figure 5. Coimmunoprecipitation analysis of the potential associations between P48 and proteins required for intranuclear microvesicle formation. Sf9 cells were cotransfected twice with transient-expression plasmids (A) or coinfected with recombinant viruses (B) separately expressing P48:FLAG and Myc-tagged Ac75, Ac76, Ac93, or P48. At 36 h p.t. or 36 h p.i., the cells were harvested, and the proteins were immunoprecipitated with protein A/G agarose beads conjugated to an anti-Myc antibody. Western blot analyses were performed with an anti-FLAG antibody to detect the expression (Input, top panels) and coimmunoprecipitation (IP: anti-Myc, top panels) of FLAG-tagged P48 and with an anti-Myc antibody to detect the expression (Input, bottom panels) and immunoprecipitation (IP: anti-Myc, bottom panels) of Myc-tagged viral proteins. Cells cotransfected with pIB-P48:FLAG and pIB-V5/His or cells coinfected with vAcWT-P48:FLAG and vAcWT were used as negative controls. Input, cell lysates; IP: anti-Myc, immunoprecipitation with protein A/G agarose beads conjugated to an anti-Myc antibody. IgH, immunoglobulin heavy chain. IgL, immunoglobulin light chain. The molecular masses (in kDa) of protein standards are noted on the left.
To further investigate whether P48 associates with Ac75, Ac76, or P48 itself in the presence of viral infection, coimmunoprecipitation assays were performed on virus-infected cells. For this assay, recombinant wild-type viruses that expressed FLAG-tagged P48 or Myc-tagged Ac75, Ac76, Ac93, or P48 were constructed. Sf9 cells coinfected with vAcWT-P48:FLAG and vAcWT-Ac75:Myc, vAcWT-Ac76:Myc, vAcWT-Ac93:Myc, or vAcWT-P48:Myc were harvested at 36 h p.i., and the cell lysates were subjected to immunoprecipitation with protein A/G agarose beads conjugated to an anti-Myc antibody. Cells coinfected with vAcWT-P48:FLAG and vAcWT were used as a negative control. The expression of FLAG-tagged and Myc-tagged proteins was confirmed byWestern blotting with an anti-FLAG or anti-Myc antibody (Fig. 5B, Input lanes). As shown in Fig. 5B, the association between P48 and Ac93 was confirmed, but the association of P48 with Ac76, Ac75, and itself was not detected.
Taken together, the results described above indicated that P48 associates with Ac93 in the absence of viral infection, and there may be no association between P48 and Ac75, Ac76, or P48 itself or that the associations may be too weak to be detected, even in virus-infected cells.
p48 Is Required for the Efficient Formation of Intranuclear Microvesicles
P48 Is Localized to the Nucleocapsids and Envelopes of Both BVs and ODVs
P48 Is Localized to the Plasma Membrane, Nuclear Envelope, IntranuclearMicrovesicles, andtheEnvelope and Nucleocapsid of ODVs in Infected Cells
P48 Associates with Ac93
-
p48 has been reported to be required for ODV formation (Yuan et al. 2008); however, the precise function of p48 during ODV morphogenesis, particularly whether the lack of ODV production in p48-null virus was attributed to a defect in intranuclear microvesicle formation or the failure of nucleocapsid envelopment at intranuclear microvesicles, remains to be addressed. In this study, we demonstrated that p48 is required for the efficient formation of intranuclear microvesicles. P48 is a structural protein of the nucleocapsid and envelope of both BVs and ODVs. This protein associates with another viral structural protein, Ac93, which is required for intranuclear microvesicle formation.
Western blot analysis showed that P48 was present in both BVs and ODVs (Fig. 3). However, this protein was not included in the list of BV- or ODV-associated proteins identified by proteomic studies (Braunagel et al. 2003; Wang et al. 2010). It is possible that the amount of P48 associated with BVs and ODVs was too low to be detected or that P48 is not amenable to be detected by proteomic techniques used in these studies. Further analysis by viral fractionation showed that P48 was associated with both the envelope and nucleocapsid of BVs and ODVs (Fig. 3). The distribution pattern of P48 is different from that of Ha91, the homolog of P48 in Helicoverpa armigera NPV (HearNPV), which was identified by proteomic analysis in both the nucleocapsid and envelope fractions of BVs but not as a component of ODVs (Hou et al. 2013). This may be attributed to the divergence in various virus species. It is not unusual for baculovirus proteins to present different distribution patterns in virions between homologs. For example, AcMNPV P33 (Ac92) is associated with the envelope of BVs and the nucleocapsid and envelope of ODVs (Wu and Passarelli 2010), while its homolog in Bombyx mori NPV (BmNPV) was not detected in either BVs or ODVs (Ge et al. 2011), and its homolog in HearNPV was only detected in the nucleocapsid and envelope fractions of ODVs (Hou et al. 2013). AcMNPV VP80 (Ac104) is located to the nucleocapsid of BVs and ODVs (Marek et al. 2011), while its homolog in HearNPV is located to nucleocapsid of BVs and both nucleocapsid and envelope of ODVs (Hou et al. 2013). AcMNPV P24 (Ac129) and its homolog in OpMNPV are located to both BVs and ODVs (Wolgamot et al. 1993), while its homolog in Spodoptera litura NPV (SpliNPV) is only associated with ODVs (Li et al. 2005b). Additionaly, AcMNPV Ac66 and Ac132 are located to BVs (Ke et al. 2008; Yang et al. 2014), while their homologs in BmNPV were not found to be associated with BVs (Zhou et al. 2010).
Immunogold-labeled P48 was localized to the plasma membrane, nuclear envelope, intranuclear microvesicles, nucleocapsids in the VS, and the envelope and nucleocapsid of ODVs in virus-infected cells (Fig. 4), which coincides with the localization pattern of P48 on purified virions (Fig. 3). The distribution of P48 to the plasma membrane, nuclear envelope and intranuclear microvesicle is consistent with its association with envelope fraction of both BVs and ODVs, and the localization of P48 on the nucleocapsids in virus-infected cells makes it reasonable that P48 is present in the nucleocapsid fraction of both BVs and ODVs. The association of P48 with various structures of virus-infected cells and virions reflected that this protein might play multiple functions in baculovirus life cycle. In addition to the involvement in intranuclear microvesicle formation as described above and its critical role in nuclear egress of nucleocapsids as previous described (Yuan et al. 2008; Yue et al. 2018), which coincides with the distribution of P48 at the nuclear envelope, the localization of P48 to the plasma membrane and BV envelope suggested that this protein might be involved in the assembly of BV at the plasma membrane. Besides the membrane or envelope structure, P48 is also targeted to nucleocapsids. As the deletion of p48 does not seem to affect the assembly of nucleocapsids and transport of nucleocapsids from the VS to the ring zone (Yuan et al. 2008), it is possible that P48 in the nucleocapsid may be involved in the recognition and interaction with the viral protein-modified nuclear membrane, plasma membrane, or intranuclear microvesicles to facilitate nuclear egress, or envelopment of BVs or ODVs. In order to develop a comprehensive understanding of the functional role of P48 in viral life cycle, further studies are necessary to test these hypotheses.
Electron microscopy analysis showed that deletion of p48 led to a defect in intranuclear microvesicle formation. Besides p48, three baculovirus genes including ac76, ac93, and ac75 have been reported to be involved in intranuclear microvesicle formation. Thus, the association of P48 with the above gene products was investigated. The result showed that P48 associated with Ac93. No association was detected between P48 and Ac75, Ac76, or P48 itself in this study. This result is slightly different from that of a recent study which showed that P48 was associated with Ac76 (Yue et al. 2018). It may be because the association between P48 and Ac76 is quite weak and may not be easily detected in coimmunoprecipitation assay, as P48 and Ac76 showed a weaker fluorescence complementation signal than other combinations by bimolecular fluorescence complementation analysis (Yue et al. 2018). Association between Ac93 and Ac76 was also detected by Yue et al. (2018). In addition, our previous study showed that Ac75 interacts with Ac76. Whether P48, Ac75, Ac76 and Ac93 form a complex to play roles in intranuclear microvesicle formation and the exact interaction network among these proteins need further works to be revealed. In addition to the trafficking of ODV membrane proteins to modify the nuclear membrane, the formation of intranuclear microvesicles requires the budding of the nuclear membrane (Braunagel and Summers 2007; Hellberg et al. 2016). It is necessary to figure out the exact step of intranuclear microvesicle formation these proteins function in future investigations.
-
We thank Prof. Zhihong Hu (Wuhan Institute of Virology) for the generous gift of E25 polyclonal antiserum. This research was supported by the National Natural Science Foundation of China (31572056 and 31872025), the Key Project of Natural Science Foundation of Guangdong Province (2018B030311018), the National Key R & D Program of China (2017YFD0200404), and the Guangzhou Science and Technology Project (201707020003).
-
YW and MY conceived the experiments. YW constructed recombinant viruses and conducted TEM analyses, TCID50 EPDA, BV and ODV purification, IEM analyses, and coimmunoprecipitation assays. QC and JC constructed plasmids. ZH, WW, KY and MY analyzed the results. MY is responsible for the financial support of the project. All authors reviewed the manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: