HTML
-
SAM domain and HD domain-containing protein 1 (SAMHD1) is an antiviral dNTP triphosphohydrolase (dNTPase) that suppresses lentiviral replication by reducing the concentration of cellular dNTPs to a level insufficient for viral cDNA synthesis (Goldstone et al. 2011; Powell et al. 2011; Kim et al. 2012; Lahouassa et al. 2012; St Gelais et al. 2012). However, lowering cellular dNTP levels may not be the sole mechanism of restriction (White et al. 2013b; Bhattacharya et al. 2016; Welbourn and Strebel 2016). Activation of the dNTPase activity of SAMHD1 relies on its catalytically-active tetrameric state initiated by GTP/dGTP binding (Amie et al. 2013; Ji et al. 2013, 2014; Koharudin et al. 2014; Zhu et al. 2015). Phosphorylation of SAMHD1 by cyclin-dependent kinases at the Thr 592 residue reduces the stability of the SAMHD1 tetramer, leading to the loss of antiviral activity of SAMHD1 (Cribier et al. 2013; White et al. 2013b; Pauls et al. 2014; Tang et al. 2015; Yan et al. 2015). As counter mechanisms, viral protein X (Vpx) from human immunodeficiency virus type 2 (HIV-2) and some strains of simian immunodeficiency virus (SIV) recruits SAMHD1 to the E3 ubiquitin ligase complex CRL4DCAF1 for ubiquitination (Srivastava et al. 2008; Bergamaschi et al. 2009; Hrecka et al. 2011; Laguette et al. 2011; Ahn et al. 2012). This recruitment leads to proteasomal degradation of SAMHD1, which elevates cellular dNTP concentration and, therefore, relieves SAMHD1-mediated restriction of lentiviral replication in monocyte/macrophages and resting T cells (Hrecka et al. 2011; Laguette et al. 2011; Ahn et al. 2012; Baldauf et al. 2012; Berger et al. 2012; Descours et al. 2012). HIV-1 lacks a Vpx protein and its viral protein R (Vpr) cannot mediate the proteasomal degradation of SAMHD1, although some other SIV strains employ the related Vpr protein to fulfill the same function (Lim et al. 2012; Spragg and Emerman 2013). In addition to primate lentiviruses, SAMHD1 also has been shown to restrict a variety of other retroviruses and DNA and other RNA viruses, such as feline immunodeficiency virus (FIV), bovine immunodeficiency virus (BIV), hepatitis B virus and enterovirus 71 (Gramberg et al. 2013; Hollenbaugh et al. 2013; Kim et al. 2013; White et al. 2013a; Chen et al. 2014; Li et al. 2020).
Wild-type (WT) human SAMHD1 (hSAM) is predominantly localized to the nucleus, which is mediated by a nuclear localization signal (NLS) in the N-terminus of SAMHD1. Disruption of the NLS results in cytoplasmic accumulation of the SAMHD1 protein and renders it resistant to Vpx-induced degradation (Brandariz-Nunez et al. 2012; Hofmann et al. 2012; Wei et al. 2012). However, cytoplasmic hSAM still interacts with Vpx, maintains dNTPase activity and inhibits lentiviral infection, suggesting that the intracellular dNTPs can freely diffuse throughout the cell (Brandariz-Nunez et al. 2012; Hofmann et al. 2012). Many SAMHD1 mutations found in patients with Aicardi-Goutières syndrome (AGS) can also disrupt nuclear localization, leading to SAMHD1 accumulation in the cytoplasm (Rice et al. 2009; Goncalves et al. 2012; White et al. 2017).
Feline SAMHD1 (fSAM) and bovine SAMHD1 (bSAM) could restrict their host species-carrying lentiviruses FIV and BIV, respectively, by using their dNTPase activities (Mereby et al. 2018). To facilitate the establishment of alternative animal models in addition to mice for investigating SAMHD1 in vivo, we previously studied the restriction profile of fSAM and bSAM against different primate lentiviruses and the molecular mechanisms of fSAM- and bSAM-mediated restriction (Wang et al. 2020). However, the intracellular distribution of fSAM and bSAM is not very clear, and the roles of intracellular distribution of fSAM and bSAM in cellular physiology and viral restriction are not well understood.
In this study, we first confirmed that the NLS sequence 11KRPR14 was conserved in fSAM and bSAM and found that the NLS-deleted (NLS) fSAM and bSAM relocalized to the cytoplasm. Then we investigated the antiviral function of cytoplasmic fSAM and bSAM against different lentiviruses and found that cytoplasmic fSAM could restrict SIVmac239 and HIV-2 ROD more efficiently than its WT protein by being less sensitive to Vpx-mediated degradation like cytoplasmic hSAM. While cytoplasmic bSAM was different from cytoplasmic hSAM and fSAM in sensitivity to Vpx-mediated degradation, although it still retained the antiviral activity against different lentiviruses. These findings suggested that fSAM- and bSAM-mediated lentiviral restriction does not require their nuclear localization and that feline SAMHD1 shares more common features with human SAMHD1, thus is more suitable for the establishment of the animal model to study SAMHD1 in vivo.
-
HEK293, HEK293T and CrFK cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). TZM-bl and U937 cells were provided by the National Institutes of Health-AIDS Reagent Program (NIH-ARP). HEK293, HEK293T, TZM-bl and CrFK cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Stable U937 cells were maintained in RPMI 1640 medium with 10% FBS and 1 μg/mL puromycin and were differentiated with PMA (phorbol 12-myristate 13-acetate; 25 ng/mL) for 20 h before use. All cell lines were cultured at 37 ℃ and 5% CO2.
-
Plasmids pVR1012-homo-SAMHD1-HA, pCG-Vpxmac-HA, and pCG-Vpxmac/Q76A-HA were gifts from Dr. Wenyan Zhang (Institute of Virology and AIDS Research, First Hospital of Jilin University). pVR1012-feline-SAMHD1-HA, pVR1012-bovine-SAMHD1-HA, pVR1012-Vpxmac-myc, pVR1012-VpxROD-myc and pVR1012-VpxROD/Q76A-myc were constructed as previously described (Wang et al. 2020). NLS-deleted human, feline and bovine SAMHD1 fragments with C-terminal hemagglutinin (HA) tags were generated by PCR using pVR1012-homo-SAMHD1-HA, pVR1012-feline-SAMHD1-HA and pVR1012-bovine- SAMHD1-HA as templates, respectively. The 11-14 amino acids KRPR at the N-terminus of each species of SAMHD1 were deleted by using the following primers: for hSAMΔNLS, forward primer: 5'-GCTCTAGAATGCAG CGAGCCGATTCCGAGCAGCCCTCCTGCGATGACA- 3'; for fSAMΔNLS, forward primer: 5'-GCTCTAGAATG CAGGGAGCAGACTCAGACCAGCCCTTCCGCGATG GCA-3'; for bSAMΔNLS, forward primer: 5'-AAGTCTAGAATGCAGAGTGCCGACTCCCAGAACACCCCC CGCGATGGCAGCCCAAGAAC-3'; the reverse primer for all three species of SAMHD1 was: 5'-CGGGATCCTTACGCGTAATCTGGGACGTCGTAAGGG-3'. All PCR products were digested and cloned into the XbaI and BamHI sites of pVR1012 vector. To construct enhanced green fluorescent protein (EGFP)-fused SAMHD1 proteins, the EGFP fragment was amplified from a plasmid pEGFPN1 (Clontech, Mountain View, CA, USA) by using the forward primer: 5'-ACGCGTCGACATGGTGAGCAAGG GCG-3' and reverse primer: 5'-GCTCTAGACTTGTACAGCTCGTCCATGC-3'. The PCR product was inserted into the WT and NLS-deleted SAMHD1-expressing plasmids at the SalI and XbaI restriction sites. A plasmid pVR1012-EGFP expressing EGFP only was also constructed as a systemic control. HIV-2 ROD Vpx-expressing lentiviral backbone plasmid pLVX-VpxROD was constructed by inserting the PCR-amplified Vpx fragment with a C-terminal FLAG tag into the XhoI and BamHI sites of pLVX-puro vector.
-
Anti-HA mouse monoclonal antibody (mAb) (no. 901514) was purchased from Covance (Princeton, NJ, USA). Antimyc mouse mAb (no. 05-419) was purchased from Millipore (Burlington, MA, USA). Anti-GAPDH mouse mAb (no. 60004-1-Ig) and anti-histone H3 mAb (no. 17168-1- AP) were purchased from Proteintech Group (Rosemont, IL, USA). Anti-SIV p27 mAb was provided by NIH-ARP. The proteasome inhibitor MG132 in DMSO solution (no. M7449) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Alkaline phosphatase-conjugated goat antimouse IgG was obtained from Jackson Immunoresearch (Baltimore Pike, PA, USA).
-
DNA transfection was carried out using jetPRIME transfection reagent (Polyplus transfection, Illkirch, France) according to the manufacturer's instructions. In co-transfection assays, cells were harvested at 48 h post-transfection, centrifuged at 3000 ×g for 5 min and lysed with RIPA buffer (pH 7.4, containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 1 mmol/L EDTA). For preparation of nuclear and cytoplasmic extracts, cell pellets harvested from 12-well plates were first resuspended in 80 μL of RLN buffer (pH 7.4, containing 50 mmol/L Tris- HCl, 40 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.5% Nonidet P-40) in the presence of protease inhibitor (Roche, South San Francisco, CA, USA) and incubated on ice for 5 min, and then centrifuged at 3000×g for 10 min at 4 ℃ to extract the supernatant as the cytoplasmic samples. The nuclear pellet was washed once with the RLN buffer and then resuspended in 20 μL of RIPA buffer. After vortexed for 30 min, the nuclear samples were centrifuged at 12,000 ×g for 5 min at 4 ℃ to remove debris. In the SAMHD1 degradation assay with lentiviruses carrying Vpx, Lenti-VpxROD pseudoviruses were produced by transfection into HEK293T cells with pLVX-VpxROD, psPAX2 (Addgene, Watertown, MA, USA) and pVSV-G at a mass ratio of 1.6: 2: 1. The titer of Lenti-VpxROD was determined by the concentration of p24 (Alliance HIV-1 P24 ANTIGEN ELISA Kit, PerkinElmer, Waltham, MA, USA). HEK293 cells (5 × 105) were infected with 3 lg p24 of Lenti-VpxROD or incubated with DMEM supplemented with 10% FBS for 24 h, then transfected with 600 ng of different SAMHD1-HA expression plasmids or empty vector. Cells were harvested at 48 h post-transfection. All samples were mixed with 4× loading buffer and boiled at 97 ℃ for 10 min. The prepared protein samples were separated by electrophoresis on SDS-polyacrylamide gels (SDS-PAGE) and detected by immunoblotting as previously described (Wang et al. 2020).
-
HEK293 cells were seeded onto coverslips at 5 × 104 cells per well in 24-well plates one day before transfection. Cell monolayers were about 10%-20% confluence on the day of transfection. At 48 h post-transfection, monolayers were washed twice with phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 15 min. Paraformaldehyde-fixed cells were then washed twice with PBS. Cellular nuclei were labeled with 1 μg/mL of DAPI (4', 6-diamidino-2-phenylindole) in PBS for 5 min, then washed twice with PBS. Images were captured with a Confocal Laser Scanning Microscope (Zeiss, Oberkochen, Germany) using a 40× objective. For quantification of the percentage of nuclear SAMHD1 in WT and NLS-deleted SAMHD1 expressing cells, the average mean fluorescence intensity (MFI) per pixel of an area of interest in the nuclear and cytoplasmic areas were first determined using Adobe Photoshop (San Jose, CA, USA). Next, relative nuclear and cytoplasmic MFI was derived by subtracting the average background MFI from the average nuclear and cytoplasmic MFI, respectively, and sum of the relative nuclear MFI and relative cytoplasmic MFI was calculated to represent the whole cell-associated SAMHD1. The percentage of nuclear SAMHD1 was then calculated as the ratio of the relative nuclear MFI to the sum MFI.
-
Viruses were generated by transfection into HEK293T cells. The infectious molecular clones for the generation of HIV-1 (NL4-3), SIVmac239 and HIV-2 ROD were provided by NIH-ARP. The FIV pseudovirus clone (pFP93 and pGINSIN) was described previously (Khare et al. 2008; Gramberg et al. 2013). The FIV-GFP reporter virus was produced by transfection with pFP93, pGINSIN and pVSV-G at a mass ratio of 3: 3: 1. The infectious titer of HIV-1, SIVmac239 and HIV-2 ROD was determined by the concentration of reverse transcriptase (RT) using a Lenti RT Activity Kit (Cavidi, Uppsala, Sweden). The titer of FIV-GFP was determined using HEK293T cells as the number of GFP-positive cells per mL of virus stock. The viral infectivity assay using TZM-bl cells was performed as previously described (Wang et al. 2020). Briefly, 2 × 105 TZM-bl cells transfected with SAMHD1 were infected with the RT concentration-determined viruses (0.1 ng RT of HIV-1, 1 ng RT of SIVmac239 or 1 ng RT of HIV-2 ROD) at 48 h post-transfection. The cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-b-dgalactopyranoside) at 48 h post-infection. The viral infectivity was determined by the number of positive blue cells. For the viral infectivity assay using HEK293T cells, cells transfected with or without SAMHD1 were detached with trypsin at 48 h post-transfection and were re-seeded into 24-well plates (2 × 105 cells per well) with FIV-GFP at multiplicities of infection (MOI) = 0.5. At 48 h postinfection, the cells were first observed under fluorescence microscope for GFP expression and then analyzed for the percentage of GFP-positive cells by flow cytometry. Each experiment was performed in triplicate.
-
Stable U937 cells were generated and infected as previously described (Wang et al. 2020). Briefly, PMAdifferentiated SAMHD1-expressing stable U937 cells (5 × 105) were infected with HIV-1 of 0.1 ng RT, SIVmac239 of 1 ng RT or HIV-2 ROD of 1 ng RT for 4 h, then washed three times with RPMI 1640 medium and cultured for 5 days to monitor the viral replication. The culture supernatants were harvested regularly to measure the concentration of p24 for HIV-1 or RT for SIVmac239 and HIV-2 ROD. For FIV-GFP, the PMA-differentiated cells (5 × 105) were infected with different amounts of the virus (MOI = 0.5, 1.0 or 2.0) and then analyzed for the percentage of GFP-positive cells at 48 h post-infection by flow cytometry. All infections were performed in triplicate.
-
The in vitro dNTPase activity assay was performed as previously described (Wang et al. 2020). Briefly, SAMHD1-transfected HEK293 cells in 12-well plates were harvested at 48 h post-transfection, washed twice with cold reaction buffer (pH 7.4, containing 50 mmol/L Tris-HCl, 50 mmol/L KCl and 5 mmol/L MgCl2), and processed as described in the co-immunoprecipitation assay below. After incubated with anti-HA antibody-conjugated agarose beads (Roche) for 3 h, the beads were washed three times with washing buffer, once with reaction buffer and then resuspended in 40 μL reaction buffer. Partial bead slurry was reserved as the input control. The rest was diluted and incubated with 1 mmol/L dGTP and 0.01 U inorganic pyrophosphatase (New England Biolabs, Ipswich, MA, USA) at 37 ℃ for 2 h with occasional mixing. The reactions were stopped by heating to 70 ℃ for 5 min. The inorganic phosphate release from the reactions was measured using a Malachite Green Detection Kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions. All reactions were performed in triplicate.
-
Cells in 6-well plates were harvested at 48 h posttransfection, washed twice with cold PBS and lysed in lysis buffer (pH 7.4, containing 50 mmol/L Tris-HCl, 150 mmol/L NaCl and 1% Triton X-100) supplemented with the protease inhibitor at 4 ℃ for 40 min. The cell lysates were centrifuged at 16,000×g for 15 min and the supernatants were incubated with anti-HA beads at 4 ℃ for 3 h. Then, the beads were washed three times with washing buffer (pH 7.4, containing 20 mmol/L Tris-HCl, 100 mmol/L NaCl and 0.05% Tween-20), resuspended with 2 × SDS sample buffer and boiled at 97 ℃ for 10 min. The prepared samples were subjected to SDS-PAGE and Immunoblotting.
-
Data are shown as mean ± standard deviation (SD). Significance is calculated by using unpaired Student's t-test or repeated-measure analysis of two-way ANOVA with PRISM v6 (GraphPad Software, Inc., La Jolla, CA, USA). In all figures, * indicates P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; ns indicates no significance.
Cell Lines and Cell Culture
Plasmids
Antibodies and Chemicals
Western Blotting
Immunofluorescence Microscopy
Viral Infectivity Assay
Generation and Infection of Stable U937 Cell Lines
In vitro dNTPase Activity Assay
Co-immunoprecipitation Assay
Statistical Analysis
-
Previous works have demonstrated that the NLS 11KRPR14 of human SAMHD1 is required for its nuclear localization (Brandariz-Nunez et al. 2012; Hofmann et al. 2012). Alignment of amino acid sequences of the N-terminal domain of different SAMHD1 proteins demonstrated that 11KRPR14 is conserved in the human, simian, feline and bovine homologues (Fig. 1A). To determine whether the 11KRPR14 motif is also essential to the nuclear localization of fSAM and bSAM, we generated expression vectors for the NLS-deleted forms of human, feline and bovine SAMHD1 (hSAMΔNLS, fSAMΔNLS and bSAMΔNLS) with HA tags, then fused EGFP to N-terminus of the WT or NLS-deleted SAMHD1 proteins, and tested the different constructs for expression and SAMHD1 intracellular distribution in HEK293 cells. The expression of EGFP-fused SAMHD1 proteins in the transfected HEK293 cells was first analyzed by Western blotting (Fig. 1B). Next, the intracellular distribution of these SAMHD1 proteins in HEK293 cells was analyzed by immunofluorescence experiments. Compared to the intracellular distribution of EGFP expressed by a control vector pVR1012-EGFP in both the nucleus and cytoplasm, all WT SAMHD1 proteins were localized to the nucleus of HEK293 cells (Fig. 1C). By contrast, all SAMHD1 variants missing the NLS were localized to the cytoplasm, indicating that the 11KRPR14 sequence also acts as the nuclear localization signal of fSAM and bSAM. Further quantification of the percentage of nuclear SAMHD1 in the WT and NLS-deleted SAMHD1-expressing cells demonstrated that averagely 94.8% and 15.2% of hSAM and hSAMΔNLS, 90.6% and 30.6% of fSAM and fSAMΔNLS, and 92.2% and 18.1% of bSAM and bSAMΔNLS were localized to the nucleus across the cell population, respectively (Fig. 1D). The percentage of fSAMΔNLS in the nucleus was significantly higher than that of hSAMΔNLS. We also investigated the relative abundance of SAMHD1 protein in nuclear and cytoplasmic extracts by Western blotting (Fig. 1E). After normalized by the level of histone H3 in the nuclear extracts, the relative level of fSAMΔNLS detected in the nucleus of HEK293 cells was averagely 34.6% of its WT SAMHD1 protein in the nucleus, which was significantly higher than those percentages of hSAMΔNLS and bSAMΔNLS (17.8% and 20.8%, respectively, Fig. 1F). However, the percentages of all WT SAMHD1 proteins distributed in the cytoplasm relative to their NLS-deleted variants in the cytoplasm were similar (in a range from 16.0% to 20.6%). These results suggested that feline SAMHD1 may have an additional nucleus-targeting mechanism in addition to the classical NLS.
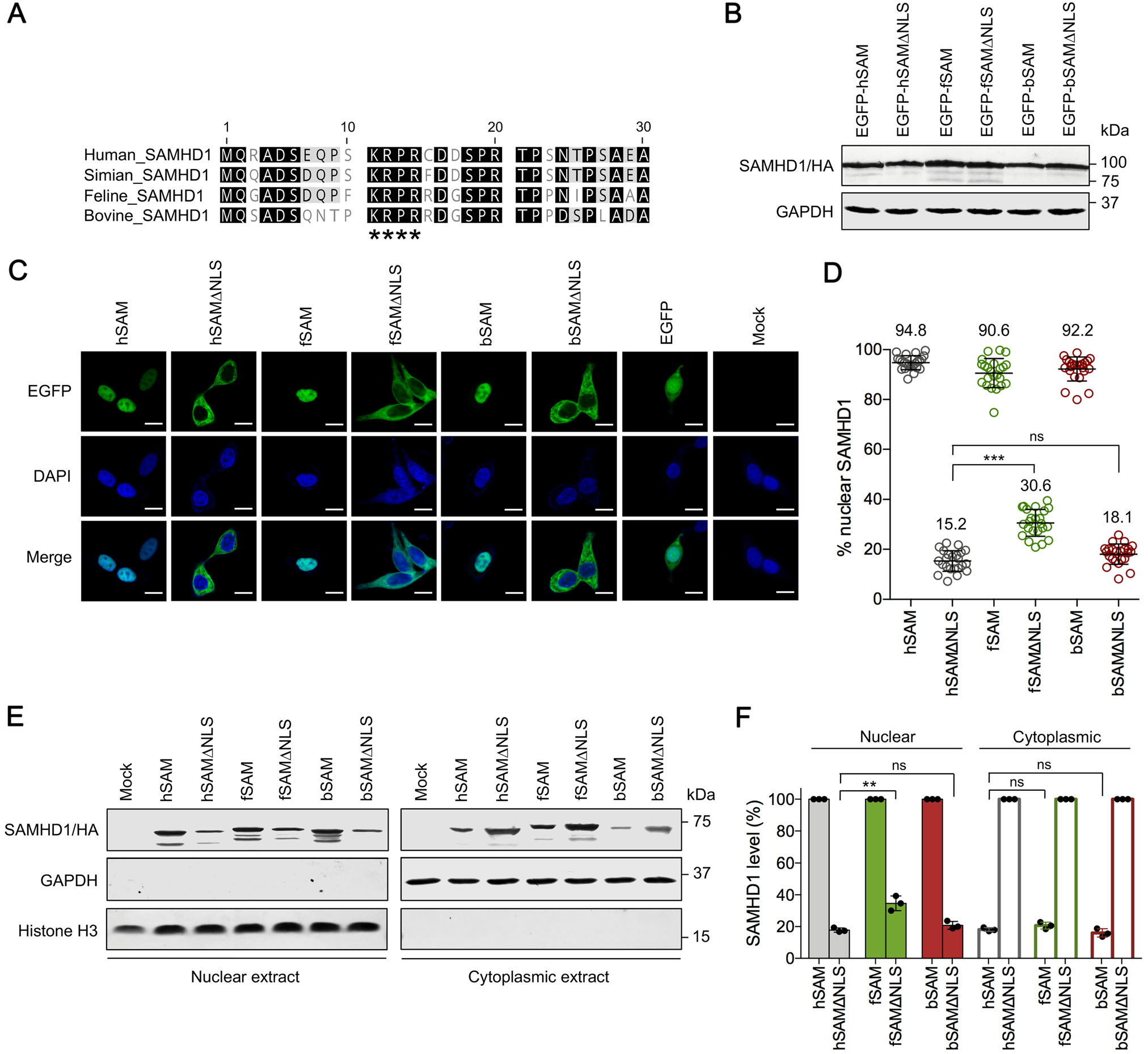
Figure 1. Identification of 11KRPR14 as the functional nuclear localization signal (NLS) of feline and bovine SAMHD1. A Alignment of the first 30 amino acids of human, simian, feline and bovine SAMHD1 proteins. The 11KRPR14 residue is indicated by asterisks. B Expression of enhanced green fluorescent protein (EGFP)-fused wild-type (WT) and NLS-deleted SAMHD1 proteins in HEK293 cells. 48 h posttransfection, the cell lysates of HEK293 cells were analyzed by Western blotting using anti-hemagglutinin (HA) antibody to detect the indicated SAMHD1 proteins with C-terminal HA-tags. GAPDH was detected as a loading control. C Intracellular distribution of WT and NLS-deleted SAMHD1 proteins in HEK293 cells. HEK293 cells expressing the indicated EGFP-fused SAMHD1 proteins (green) were fixed and detected by confocal laser scanning microscopy. Cellular nuclei were stained by DAPI (blue). White scale bars, 10 lm. D Quantification of the percentage of nuclear SAMHD1 for EGFPfused WT and NLS-deleted SAMHD1-expressing cells. Each circle represents one cell and the numbers indicate average percentages of cells shown. Statistical analysis was performed between the indicated groups using unpaired Student's t-test. E Detection of SAMHD1 by Western blotting in nuclear and cytoplasmic extracts of HEK293 cells. Samples were collected at 48 h post-transfection. Histone H3 and GAPDH were detected as nuclear and cytoplasmic protein loading controls, respectively. F Quantitation of the band intensity of nuclear and cytoplasmic SAMHD1 of the Western blotting results in HEK293 cells. SAMHD1 band intensities were quantified using Adobe Photoshop and normalized by the level of histone H3 or GAPDH. The percentage of normalized NLS-deleted SAMHD1 proteins in the nuclear extract or WT SAMHD1 proteins in the cytoplasmic extract was calculated relative to that of the corresponding WT or NLS-deleted SAMHD1 proteins (set to 100%, respectively). Error bars represent the SD calculated from three independent experiments. Statistical analysis was performed between the indicated groups using unpaired Student's t-test. **Indicates P ≤ 0.01; *** indicates P ≤ 0.001; ns indicates no significance.
-
Both nuclear and cytoplasmic forms of human SAMHD1 could restrict HIV-1 efficiently in stably-transfected U937 and SAMHD1-silenced THP-1 cell lines (Brandariz-Nunez et al. 2012). To understand how intracellular distribution influences the antiviral abilities of feline and bovine SAMHD1 and whether they have differences in restriction of different lentiviruses, we first utilized a previouslyestablished quick detection method by using TZM-bl cells (Wang et al. 2020). The cells were transfected with WT or NLS-deleted human, feline or bovine SAMHD1 expression vector or empty vector and were then infected with HIV-1, SIVmac239 or HIV-2 ROD at 48 h post-transfection. 48 h later, the infected cells were stained with the substrate of bgalactosidase (X-Gal) and the viral infectivity was determined by the number of positive blue cells. The results showed that hSAMΔNLS, fSAMΔNLS and bSAMΔNLS restricted HIV-1 as efficiently as their WT SAMHD1 proteins (Fig. 2A). hSAMΔNLS and fSAMΔNLS demonstrated significantly enhanced antiviral activities in restriction of SIVmac239 and HIV-2 ROD compared to their WT SAMHD1 proteins (Fig. 2B, 2C). However, bSAM and bSAMΔNLS restricted SIVmac239 and HIV-2 ROD similarly. We also investigated how these SAMHD1 proteins restricted FIV. Because infection of the FIV-GFP pseudovirus cannot generate the blue staining reaction in TZM-bl cells, we infected HEK293T cells with this virus, which had already been transfected with the same SAMHD1-expressing plasmids, and detected the percentage of GFP-positive cells by flow cytometry at 48 h postinfection. No difference was found in the antiviral activity of each of the three pairs of WT and NLS-deleted SAMHD1 proteins against FIV-GFP (Fig. 2D and Supplementary Fig. S1). At the same time, we confirmed that the intracellular levels of the three pairs of WT and NLSdeleted SAMHD1 proteins in the transfected TZM-bl and HEK293T cells were equal before infection, to exclude the possibility of dose-related fluctuation of antiviral activity resulting from different protein expression levels (Supplementary Fig. S2A and S2B).
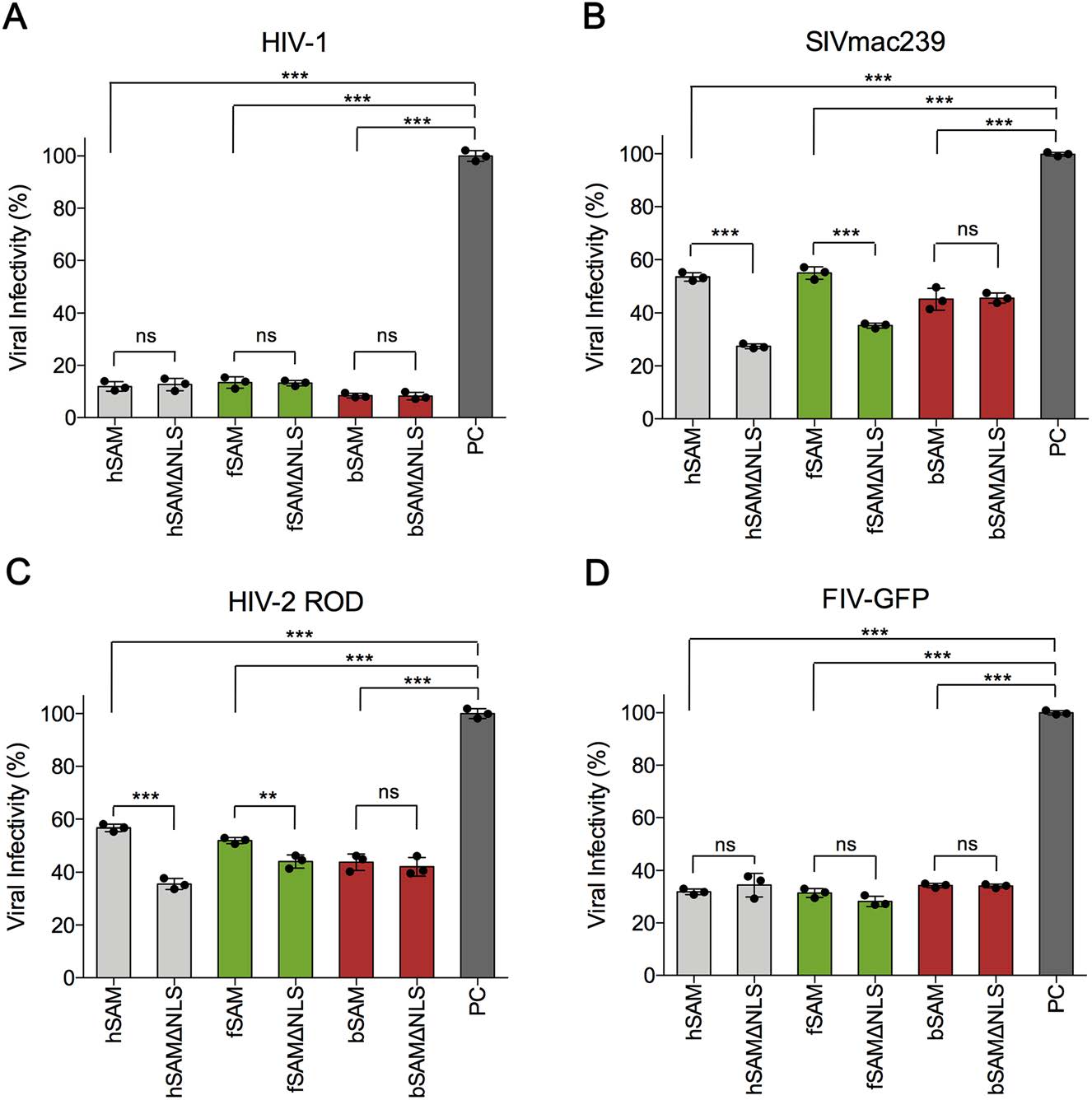
Figure 2. The antiviral activity of WT and NLS-deleted SAMHD1 proteins against different lentiviruses. A-C TZM-bl cells (2 × 106) were transfected with 1.5 μg WT or NLS-deleted human, feline or bovine SAMHD1-HA expression plasmids or empty vector and then infected with (A) HIV-1 NL4-3 (0.1 ng of reverse transcriptase, RT), (B) SIVmac239 (1 ng of RT) or (C) HIV-2 ROD (1 ng of RT) at 48 h post-transfection. Viral infectivity was determined by the number of positive blue cells stained with X-gal. D HEK293T cells (2 × 106) were transfected with 400 ng WT or NLS-deleted human, feline or bovine SAMHD1-HA expression plasmids or empty vector and then infected with FIV-GFP (multiplicities of infection, MOI = 0.5) at 48 h post-transfection. Viral infectivity was determined by the percentage of GFP-positive cells quantified by flow cytometry. The viral infectivity in empty vector-transfected cells was set to 100% (positive control, PC). Error bars represent the standard deviation (SD) calculated from three independent infections. Statistical analysis was performed between he indicated groups using unpaired Student's t-test. **Indicates P ≤ 0.01; *** indicates P ≤ 0.001; ns indicates no significance.
To further validate these results, we detected the dynamic changes of viral replication of HIV-1, SIVmac239 and HIV-2 ROD and the viral restriction effect after infection with increasing amounts of FIV-GFP in stable U937 cell lines expressing the WT or NLS-deleted SAMHD1 proteins. The results showed that all of the SAMHD1 NLS variants restricted HIV-1 and FIV-GFP as efficiently as their WT proteins (Fig. 3A, 3D). Consistent with the results obtained in TZM-bl cells, the restriction efficiency of hSAMΔNLS and fSAMΔNLS against SIVmac239 and HIV-2 ROD was significantly higher than that of their WT proteins, while the restriction efficiency of bSAM and bSAMΔNLS against these two viruses was comparable (Fig. 3B, 3C). Immunoblotting analysis of the stable U937 cells showed that the three pairs of WT and NLS-deleted SAMHD1 proteins were expressed at similar levels, respectively (Supplementary Fig. S2C). Together, these results indicated that cytoplasmic feline and bovine SAMHD1 retain the activity to inhibit the infection and replication of different lentiviruses.
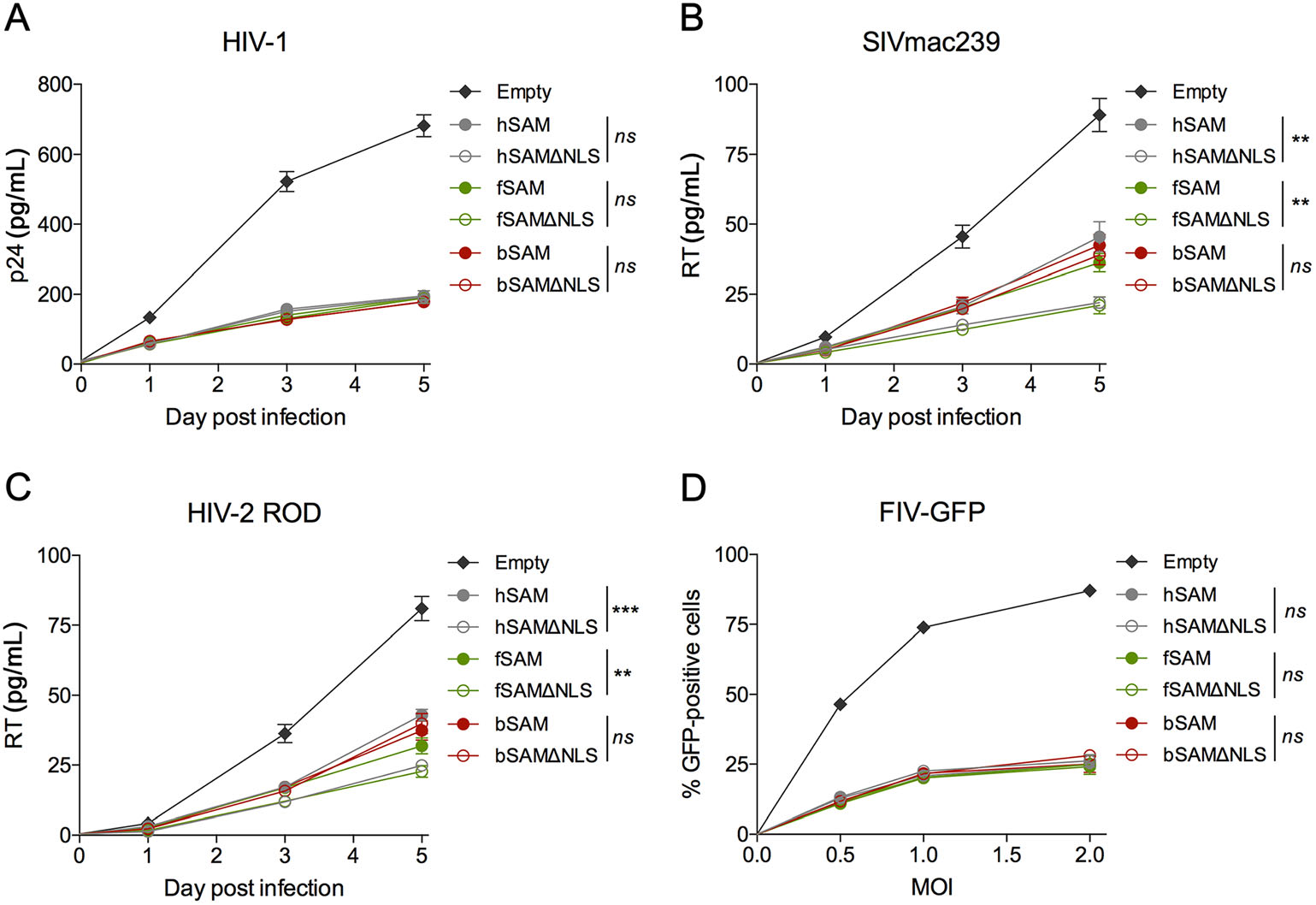
Figure 3. Restriction of viral infection and replication mediated by WT and NLS-deleted SAMHD1 proteins in U937 cells. A-D Empty U937 cells (5 × 105) or U937 cells stably expressing WT or NLS-deleted human, feline or bovine SAMHD1 protein were differentiated with PMA for 20 h and infected with (A) HIV-1 NL4-3 (0.1 ng of RT), (B) SIVmac239 (1 ng of RT), (C) HIV-2 ROD (1 ng of RT) or (D) FIV-GFP (MOI = 0.5, 1.0 and 2.0). The viral replication curves in (A-C) were measured by the concentration of p24 for HIV-1 or RT for SIV and HIV-2 in the culture supernatants collected on the day of infection (day 0) and at day 1, 3 and 5 post-infection. The percentage of GFP-positive cells in (D) was determined by flow cytometry. Error bars represent the SD calculated from three independent infections. Statistical analysis was performed between groups of each pairs of WT and NLS-deleted SAMHD1 proteins using repeated-measure analysis of two-way ANOVA. **Indicates P ≤ 0.01; *** indicates P ≤ 0.001; ns indicates no significance.
-
It is known that Vpx is encoded by HIV-2 and some strains of SIV to counteract human SAMHD1-mediated restriction by recruiting the CRL4DCAF1 E3 ubiquitin ligase complex, leading to the ubiquitination and proteasomal degradation of hSAM (Hrecka et al. 2011; Laguette et al. 2011; Ahn et al. 2012). However, cytoplasmic hSAM is less sensitive to Vpx-induced degradation (Brandariz-Nunez et al. 2012; Hofmann et al. 2012; Wei et al. 2012). Therefore, we hypothesized that the resistance of cytoplasmic SAMHD1 to Vpx-induced degradation leads to the sensitivity of SIV and HIV-2 to cytoplasmic SAMHD1-mediated restriction. Previously, we found that both WT fSAM and bSAM could be proteasomally degraded in the presence of Vpx, although they were naturally less sensitive to Vpx-mediated degradation than hSAM (Wang et al. 2020). To test whether cytoplasmic fSAM and bSAM is more resistant to Vpx-induced degradation than their WT proteins, Vpx from SIVmac239 and HIV-2 ROD strains (Vpxmac and VpxROD) and their Q76A mutants (Vpxmac/Q76A and VpxROD/Q76A), which no longer bind DCAF1 and were found unable to induce the degradation of SAMHD1 (Srivastava et al. 2008; Bergamaschi et al. 2009; Hrecka et al. 2011), were analyzed for the abilities to counteract WT or NLS-deleted human, feline and bovine SAMHD1 proteins in HEK293 cells. In agreement with previous results (Brandariz-Nunez et al. 2012; Hofmann et al. 2012), hSAMΔNLS was more resistant to both Vpxmac and VpxROD-mediated degradation than WT hSAM, and the VpxQ76A mutants no longer induced the degradation of hSAM and hSAMΔNLS (Fig. 4A, 4D, 4G, 4J). fSAMΔNLS also showed enhanced resistance to Vpxmac and VpxROD-induced degradation than its WT protein (Fig. 4B, 4E, 4H, 4K). However, bSAM and bSAMΔNLS were degraded equally in the presence of both Vpxmac and VpxROD (Fig. 4C, 4F, 4I, 4L). Although in the presence of Vpxmac/Q76A and VpxROD/Q76A, the degradation of bSAMΔNLS was not recovered, the proteasome inhibitor MG132 treatment suppressed Vpxmac and VpxROD-induced degradation of bSAM and bSAMΔNLS (Supplementary Fig. S3). Next, we repeated the immunoblotting analysis of fSAM and fSAMΔNLS in a feline kidney epithelial cell line, CrFK. Co-expression with Vpxmac or VpxROD induced less degradation of fSAMΔNLS than that of WT fSAM in this feline cell line (Fig. 5A-5D). We then examined the degradation of fSAM and fSAMΔNLS induced by SIVmac239 provirus-expressing plasmids with or without Vpx expression (pSIV or pSIVVpx) in CrFK cells. Results showed that fSAMΔNLS was about 30% less sensitive to WT SIVmac239-induced degradation than WT fSAM (Fig. 5E, 5F). We also tried to repeat these detections for bSAM and bSAMΔNLS in a cell line of their own species. However, due to the low transfection efficiency of the Madin-Darby bovine kidney (MDBK) cell line we have, we could not obtain a clearly repeated result of the degradation assay of bSAM and bSAMΔNLS in this cell line. Still, these results indicated that feline SAMHD1 degradation in response to Vpx requires nuclear localization, while bovine SAMHD1 degradation does not require it.
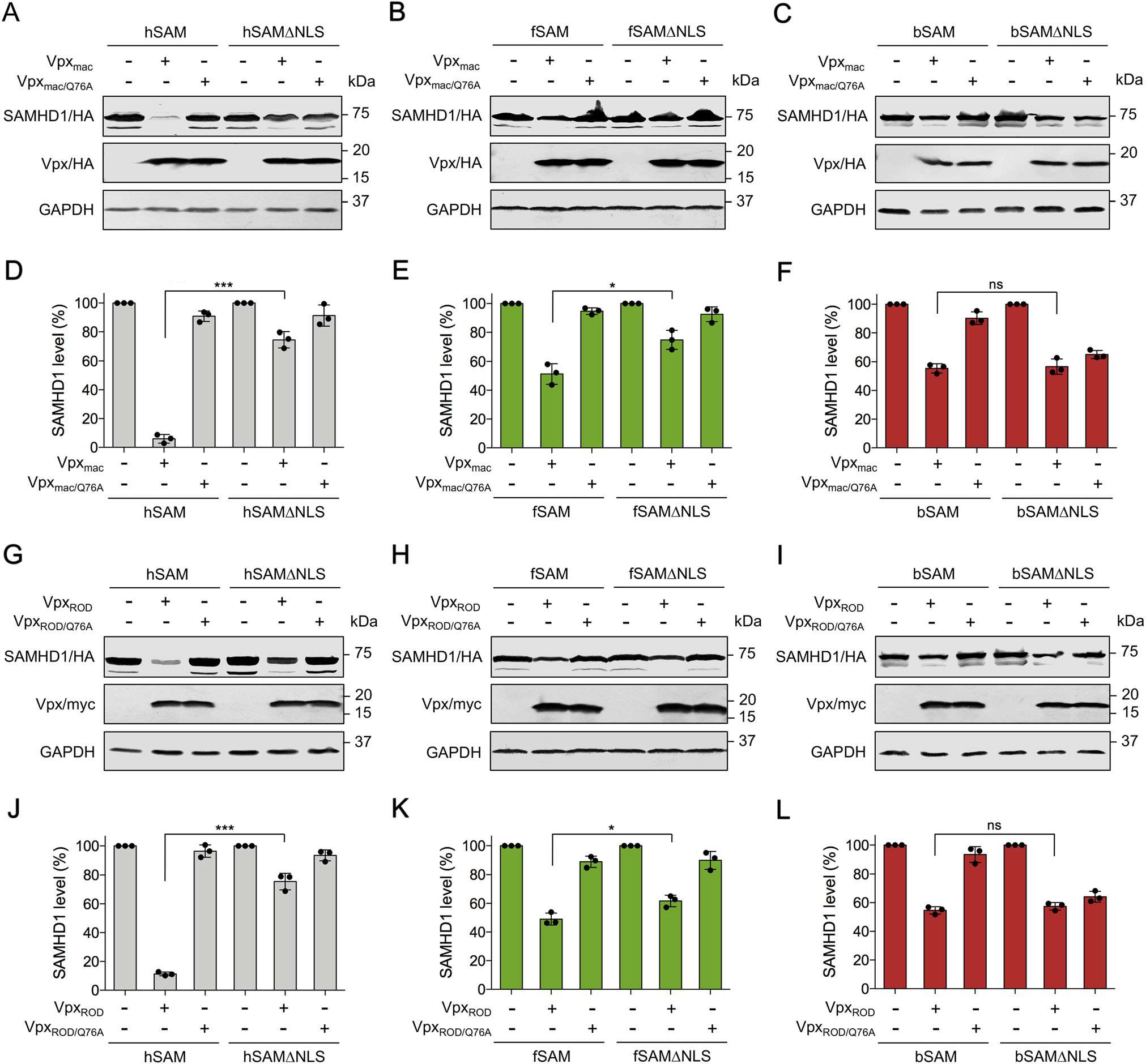
Figure 4. Cytoplasmic SAMHD1 is partially resistant to Vpx-induced degradation. A-C and G-I HEK293 cells (1 × 106) were cotransfected with 600 ng of WT or NLS-deleted (A, G) human (B, H) feline or (C, I) bovine SAMHD1-HA expression plasmids and 500 ng of pCG-Vpxmac-HA, pCG-Vpxmac/Q76A-HA, pVR1012- VpxROD-myc or pVR1012-VpxROD/Q76A-myc or empty vector. Cells were harvested at 48 h post-transfection and then analyzed by Western blotting using anti-HA, anti-myc, and anti-GAPDH antibodies. D-F and J-L Quantitation of SAMHD1 band intensity of the Western blotting results in HEK293 cells. The GAPDH-normalized percentage of SAMHD1 level in the presence of Vpxmac, VpxROD or the Vpx mutants was calculated relative to that of the corresponding empty vector-co-transfected SAMHD1 (set to 100%, respectively). Error bars represent the SD calculated from three independent experiments. Statistical analysis was performed between the indicated groups using unpaired Student's t-test. * indicates P ≤ 0.05; *** Indicates P ≤ 0.001; ns indicates no significance.
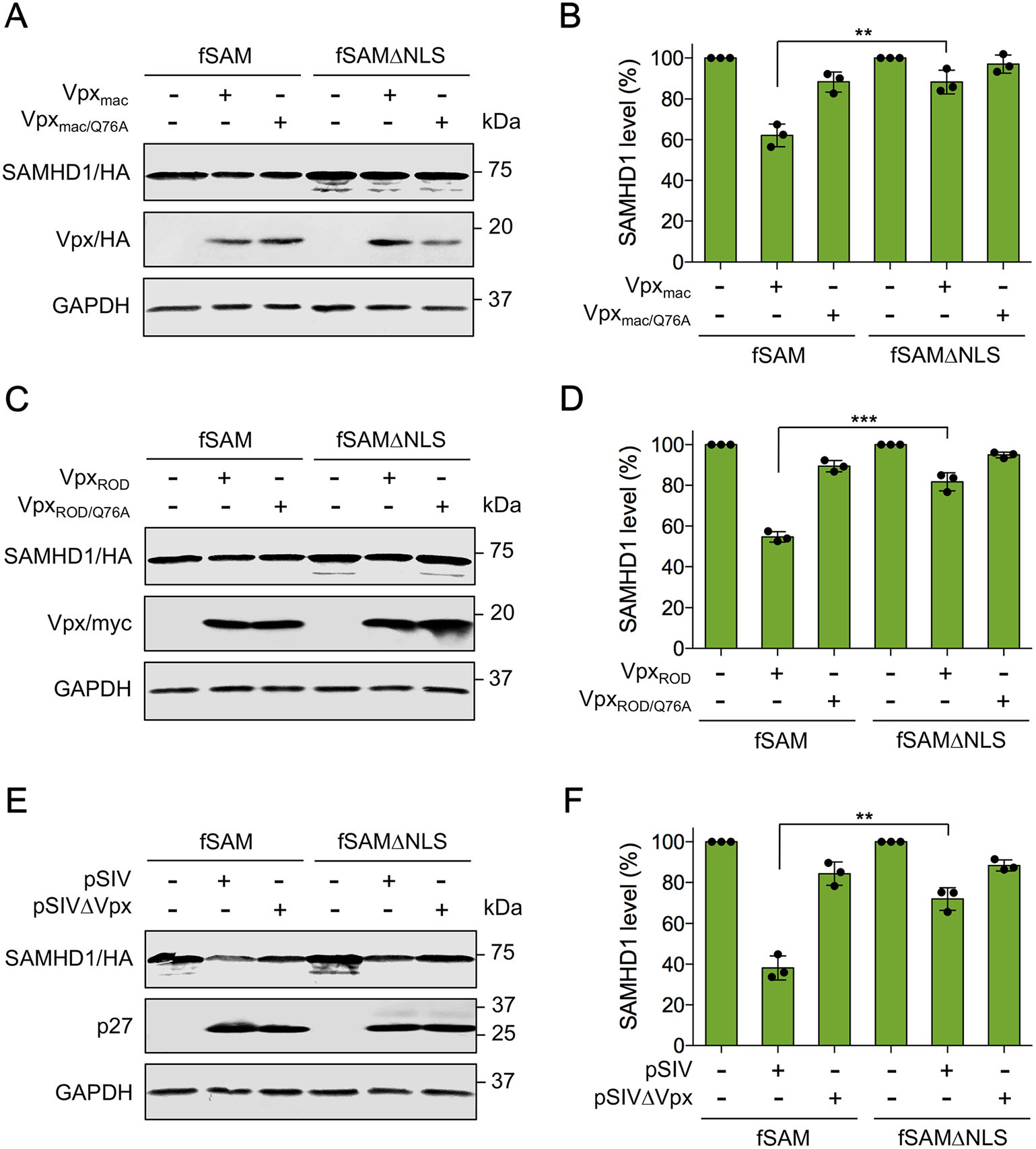
Figure 5. Vpx-induced degradation of WT and NLS-deleted feline SAMHD1 in CrFK cells. A, C, E CrFK cells (1 × 106) were cotransfected with 600 ng of WT or NLS-deleted feline SAMHD1-HA expression plasmids and 500 ng of (A) pCG-Vpxmac-HA or pVR1012-Vpxmac/Q76A-HA, (C) pCG-VpxROD-myc or pVR1012- VpxROD/Q76A-myc, or 1 μg of (E) pSIVmac239 or pSIVmac239ΔVpx, or empty vector. Cells were harvested at 48 h posttransfection and then analyzed by Western blotting using anti-HA, anti-myc, anti-p27 and anti-GAPDH antibodies. B, D, F Quantitation of SAMHD1 band intensity of the Western blotting results in HEK293 cells. The GAPDH-normalized percentage of SAMHD1 level in the presence of Vpxmac, VpxROD or SIVmac239 was calculated relative to that of the corresponding empty vector-cotransfected SAMHD1 (set to 100%, respectively). Error bars represent the SD calculated from three independent experiments. **Indicates P ≤ 0.01; ***indicates P ≤ 0.001.
-
Next, we investigated whether the phenomenon that human and feline cytoplasmic SAMHD1 are less sensitive to Vpxinduced degradation still exist when Vpx is expressed by lentivirus rather than by plasmid transfection. HEK293 cells were first infected with Lenti-VpxROD pseudoviruses carrying the vpx gene of HIV-2 ROD for 24 h, then transfected with WT or NLS-deleted human, feline and bovine SAMHD1-expressing plasmids. 48 h later, the intracellular level of SAMHD1 was determined by Western blotting. The remaining percentage of WT human, feline and bovine SAMHD1 after degradation were about 8.7%, 52.8% and 59.3%, respectively (Fig. 6A). The corresponding percentage of human and feline NLS-deleted mutants in the presence of VpxROD-carrying viruses were recovered to 40.4% and 76.1%, respectively, but the percentage of bSAMΔNLS was almost not recovered (61.5%). These results indicated that the less sensitivity of hSAMΔNLS and fSAMΔNLS to Vpx-induced degradation than their nucleus-localized WT proteins may lead to their better viral restriction outcomes.
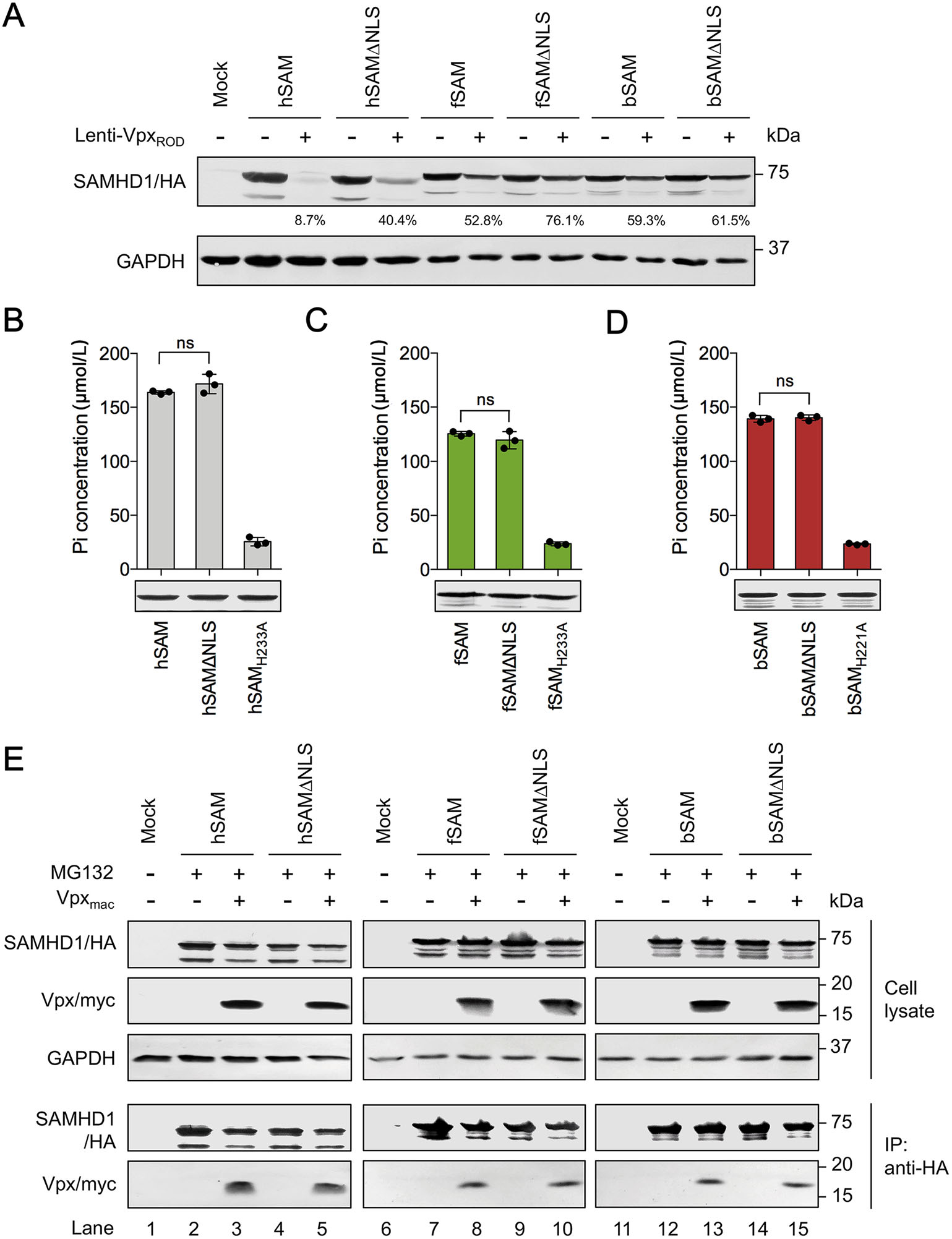
Figure 6. In vitro dNTPase activity comparison and Vpx binding capacity of WT and NLS-deleted SAMHD1 proteins. A SAMHD1 degradation mediated by Vpx-expressing lentiviruses. HEK293 cells (5 × 105) were first infected with 3 μg p24 of Lenti-VpxROD pseudovirus or incubated with DMEM supplemented with 10% FBS for 24 h, then transfected with 600 ng of WT or NLS-deleted human, feline or bovine SAMHD1-HA expression plasmids or empty vector. Cells were harvested at 48 h post-transfection and then analyzed by Western blotting using anti-HA and anti-GAPDH antibodies. The percentage of SAMHD1 in the presence of Lenti-VpxROD was calculated relative to that of the corresponding SAMHD1 in the absence of Lenti-VpxROD (set to 100%, respectively). B-D In vitro detection of SAMHD1-catalyzed inorganic phosphate (Pi) release. HA-tagged SAMHD1 proteins were isolated from transfected HEK293 cells by immunoprecipitation. An aliquot of the immunoprecipitated SAMHD1 proteins was analyzed by Western blotting to ascertain comparable protein levels using anti-HA antibody. The levels of Pi released after in vitro dGTP-pyrophosphatase hydrolysis reactions were detected by malachite green. Error bars represent the SD calculated from three independent reactions. Statistical analysis was performed between the indicated groups using unpaired Student's t-test. E Detection of the interaction between Vpx and SAMHD1 by co-immunoprecipitation. HEK293 cells (2 × 106) were co-transfected with WT or NLS-deleted human, feline or bovine SAMHD1- HA expression plasmids and 1.0 μg pVR1012-Vpxmac-myc or empty vector. All transfected cells were incubated with the proteasome inhibitor MG132 at 20 lmol/L for 12 h before harvesting. 48 h posttransfection, cell lysates were immunoprecipitated with anti-HA antibody-conjugated beads, followed by Western blotting with antiHA, anti-myc and anti-GAPDH antibodies. ns indicates no significance.
To determine whether the enhanced antiviral activity of hSAMΔNLS and fSAMΔNLS was caused by the change of the dNTPase activity due to NLS deletion, we then tested the dNTPase activity of hSAM, fSAM, bSAM and their NLS variants in vitro. Catalytically-inactive mutants hSAMH233A, fSAMH233A and bSAMH221A were also tested as the negative controls (Wang et al. 2020). The SAMHD1 proteins were immunoprecipitated with anti-HA antibodyconjugated agarose beads from transfected HEK293 cells and added to the reaction mixtures containing dGTP as the substrate and pyrophosphatase to hydrolyze the triphosphate product of SAMHD1. The final product inorganic phosphate (Pi) of hydrolysis was quantified by malachite green to determine the dNTPase activity. The results demonstrated that NLS deletion did not impact the dNTPase activity of all SAMHD1 proteins from the three species (Fig. 6B-6D).
Previous studies showed that Vpxmac and VpxROD can interact with both nuclear and cytoplasmic forms of human SAMHD1 (Hofmann et al. 2012; Schaller et al. 2014). To understand the different sensitivity of cytoplasmic fSAM and bSAM to Vpx-mediated degradation, we investigated whether cytoplasmic feline and bovine SAMHD1 still interact with Vpx. The interactions between SIVmac239 Vpx and hSAM, fSAM or bSAM or their variants with NLS deletion were analyzed by co-immunoprecipitation experiments. Myc-tagged Vpxmac and HA-tagged WT SAMHD1 or the NLS variants were co-expressed in HEK293 cells and the transfected cells were treated with MG132 for 12 h before harvest. The cell lysates were subjected to co-immunoprecipitation using anti-HA beads at 48 h post-transfection. The anti-HA beads immunoprecipitated WT SAMHD1 as well as the NLS variants from cell lysates of the transfected HEK293 cells (Fig. 6E). We observed co-precipitation of myc-tagged Vpx with all WT or NLS-deleted human, feline and bovine SAMHD1 proteins (lanes 3, 5, 8, 10, 13 and 15), indicating that all of the three cytoplasmic SAMHD1 proteins have similar Vpx binding capacity to the corresponding WT SAMHD1 proteins. Taken together with previous results, these data suggested that feline cytoplasmic SAMHD1 can suppress more SIV and HIV-2 infection and replication than its WT protein by being less sensitive to Vpx-mediated degradation as human cytoplasmic SAMHD1 rather than having higher dNTPase activity and that the less degradation of cytoplasmic human and feline SAMHD1 than their nucleus-localized WT proteins in the presence of Vpx is not caused by different Vpx binding capacity.
The NLS Sequence 11KRPR14 Is Conserved in Feline and Bovine SAMHD1
Cytoplasmic Feline and Bovine SAMHD1 Retain Antiviral Activity against Different Lentiviruses
Cytoplasmic Bovine SAMHD1 Does Not Resist Vpx-Induced Degradation
Both Nuclear and Cytoplasmic Feline and Bovine SAMHD1 Have Equal dNTPase Activity and Vpx Binding Capacity
-
To gain more knowledge of SAMHD1 from different species can provide a deeper understanding of this hostderived viral restriction factor from an evolutionary view and facilitate the establishment of animal models to understand the physiological role of SAMHD1 in vivo. SAMHD1 proteins of monkeys (Fregoso et al. 2013; Wei et al. 2014; Li et al. 2015; Schwefel et al. 2015; Shingai et al. 2015; Buchanan et al. 2016) and mice (Behrendt et al. 2013; Zhang et al. 2014; Wang et al. 2016; Bloch et al. 2017; Buzovetsky et al. 2018) have been intensively investigated for their roles in lentiviral restriction as well as for their structure, dNTPase activity and roles in the regulation of the innate immune responses. The characteristics of feline and bovine SAMHD1 have also been studied by several groups and us (Asadian et al. 2018; Mereby et al. 2018; Asadian and Bienzle 2019; Wang et al. 2020). However, the non-primate animal model of SAMHD1 has only been established in mice and knowledge of other nonprimate SAMHD1 proteins is limited. To find out more information of common and different features between the primate and non-primate SAMHD1 proteins will contribute to the improvement of SAMHD1-knockout small animal models.
A previous study of human SAMHD1 showed its distribution in both the nucleus and cytoplasm of resting CD4+ T cells, activated CD4+ T cells, and macrophages in immunoblots of nuclear and cytoplasmic fractions and 3Dreconstructed confocal images (Baldauf et al. 2012), but it is mainly localized to the nucleus when observed by confocal microscopy in Hela and U937 cells (Brandariz-Nunez et al. 2012; Hofmann et al. 2012). Study of the expression profile of SAMHD1 in feline tissues by immunohistochemical staining demonstrated that although feline SAMHD1 is predominantly localized to the nucleus, the expression level of SAMHD1 in the nucleus relative to that in the cytoplasm was variable in different tissues (Asadian et al. 2018). These results suggest that SAMHD1 is positioned into the nucleus after protein synthesis in the cytoplasm and detection of cytoplasmic SAMHD1 may reflect recently translated protein, and additionally, that SAMHD1 may be regulated by certain signals such as cytokines or phosphorylation to balance its nuclear/cytoplasmic distribution ratio in different cell lines. This can be supported by a recent study in which interferon-γ treatment was found being able to increase SAMHD1 translocation to the nucleus in primary feline CD4+ T lymphocytes (Asadian and Bienzle 2019). In the current study, we first investigated whether the NLS sequence 11KRPR14 identified in human SAMHD1 is also the key NLS in feline and bovine SAMHD1. Our quantification results of the nucleus-localized WT and NLS-deleted SAMHD1 proteins in the whole SAMHD1-expressing cells (Fig. 1D) and the immunoblotting result of the SAMHD1 proteins in nuclear and cytoplasmic fractions (Fig. 1E, 1F) demonstrated that feline NLS-deleted SAMHD1 had the highest occupation rate in the nucleus among the three NLS-deleted SAMHD1 proteins, suggesting that in addition to the N-terminal 11KRPR14 which acts as the major signal for nuclear import, feline SAMHD1 may have other signals to maintain it inside the nucleus. Some residues in feline SAMHD1 may also contribute to its nuclear localization, which is supported by the findings that mutations such as I201N, R226G and M254V of human SAMHD1 found in AGS patients were shown related to partial cytoplasmic accumulation (Goncalves et al. 2012; White et al. 2017).
Our previous study showed that feline and bovine SAMHD1 could restrict HIV-1, SIV and HIV-2 as efficiently as human SAMHD1 by using their dNTPase activities (Wang et al. 2020). Consistent with the results found in human SAMHD1 that SIVmac is not capable of overcoming the restriction imposed by the SAMHD1 NLS mutant (Brandariz-Nunez et al. 2012), hSAMΔNLS restricted more SIVmac239 and HIV-2 ROD than its WT protein in both TZM-bl and U937 cell lines (Figs. 2, 3). In our experiments, SAMHD1 was over-expressed in these two cell lines before infection and the amounts of quantified viruses used for infection were relatively low, thus, the degradation of SAMHD1 after SIV and HIV-2 infection should be incomplete. However, the difference in the antiviral activity of nuclear and cytoplasmic SAMHD1 could still be significantly seen. fSAMΔNLS showed a similar increased antiviral activity than its WT protein against SIVmac239 and HIV-2 ROD as hSAMΔNLS, suggesting that Vpx of SIVmac239 and HIV-2 ROD requires nucleus-localized human and feline SAMHD1 to induce their degradation and to overcome restriction (Fig. 7B). While bovine SAMHD1 could be degraded in both the nucleus and cytoplasm, thus the antiviral activity of bovine SAMHD1 was impaired no matter it was localized to the nucleus or the cytoplasm in the presence of Vpx (Fig. 7C). A previous study has reported that FIV and BIV infections do not induce the degradation of their host SAMHD1 proteins (Mereby et al. 2018). Therefore, all of the three pairs of nucleus- and cytoplasm-localized SAMHD1 proteins retained complete antiviral activity against HIV-1 and FIV (Fig. 7A). Considering that NLS deletion did not alter the dNTPase activity of SAMHD1 from all of the three species (Fig. 6B-6D), the better antiviral outcome was realized by the higher intracellular level of SAMHD1, confirming that SAMHD1 can deplete the overall cellular pool of dNTPs no matter in the nucleus or the cytoplasm (Brandariz-Nunez et al. 2012; Hofmann et al. 2012).
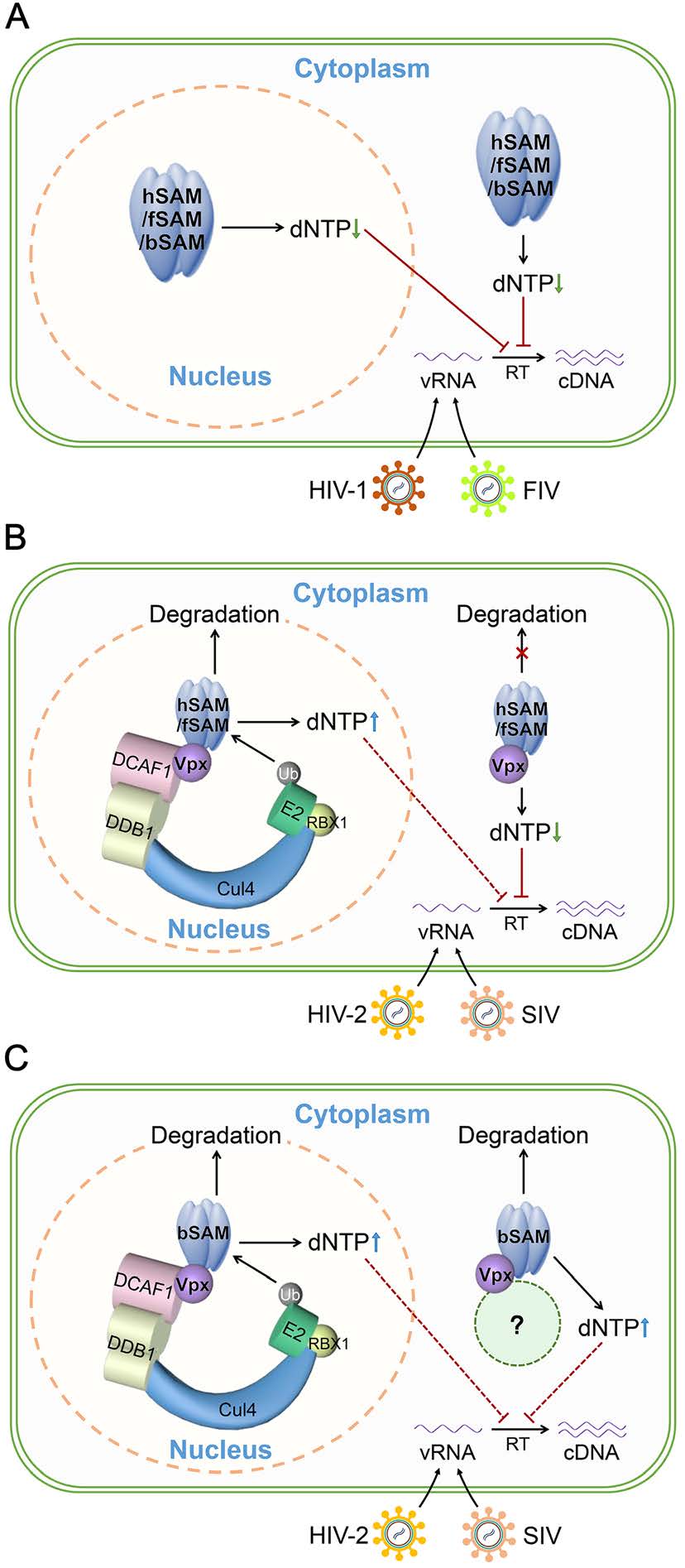
Figure 7. Proposed mechanism of feline and bovine SAMHD1-mediatedc lentiviral restriction and Vpx-mediated counteraction on different SAMHD1 proteins. A Each of tetrameric hSAM, fSAM and bSAM has dNTPase activity and both of the nucleus- and cytoplasmlocalized hSAM, fSAM and bSAM can restrict HIV-1 and FIV infection at the RT step by depleting intracellular dNTPs to a low concentration that is insufficient for cDNA synthesis from viral RNA (vRNA). B HIV-2 ROD and SIVmac239 Vpx induces degradation of nuclear hSAM or fSAM by targeting it to the CRL4DCAF1 E3 ubiquitin ligase complex and adding of ubiquitin (Ub), leading to the increase of intracellular dNTP levels and efficient viral cDNA synthesis. The red dashed line indicates the impaired viral restriction function. While cytoplasmic hSAM and fSAM are resistant to Vpxinduced degradation because Vpx cannot recruit the nucleus-localized CRL4DCAF1 E3 ubiquitin ligase complex in the cytoplasm, thus they retain the full antiviral ability. C Both nucleus- and cytoplasmlocalized bSAM can be degraded in the presence of HIV-2 ROD and SIVmac239 Vpx and their antiviral effects are also similarly impaired, thus an unknown E3 ubiquitin ligase complex (shown as the green dashed circle) which can be recruited by Vpx for specific degradation of bSAM in the cytoplasm may exist.
Previous studies suggested that Vpx-mediated degradation of human SAMHD1 is initiated in the nucleus (Brandariz-Nunez et al. 2012) and found that cytoplasmic human SAMHD1 could interact with Vpx which also has a nuclear localization signal (Pancio et al. 2000; Belshan and Ratner 2003) and retained it in the cytoplasm without being degraded (Hofmann et al. 2012). By contrast, the level of nucleus-localized human SAMHD1 is largely determined by Vpx-mediated degradation. Vpx directly binds to human SAMHD1 and simultaneously to the DCAF1 substrate receptor of the CRL4 E3 ubiquitin ligase (Hrecka et al. 2011; Ahn et al. 2012). DCAF1 was found localized in the nucleus and functioning in this compartment like other CRL4 ligases (Lee and Zhou 2007; Li et al. 2010; Guo et al. 2019). The cytoplasm-localized DCAF1 truncation mutant inhibits Vpx-mediated degradation of human SAMHD1 (Guo et al. 2019). Thus, it could be speculated that the degradation of human SAMHD1 should be confined to the nucleus. Like human SAMHD1, feline SAMHD1 with NLS deletion was less sensitive to Vpxmediated degradation than its WT protein (Figs. 4, 5), except that the recovery percentage of WT feline SAMHD1 in the presence of VpxQ76A mutants instead of WT Vpx was lower than that observed in human SAMHD1. There are two possible reasons which can be found from our results for the lower recovery percentage: (1) WT fSAM was less sensitive to Vpx-mediated degradation than WT hSAM, as found in our previous results (Wang et al. 2020); (2) NLS-deleted fSAM was not totally localized to the cytoplasm as shown in Fig. 1. Surprisingly, we found that bovine SAMHD1 was sensitive to Vpx-induced proteasomal degradation in both the nucleus and cytoplasm (Supplementary Fig. S3), but in the presence of the Q76A mutants of both SIVmac239 and HIV-2 ROD Vpx which is essential in Vpx-DCAF1 binding, the degradation of bSAMΔNLS was not recovered significantly as what was observed in hSAMΔNLS and fSAMΔNLS (Fig. 4). We further showed that both nucleus- and cytoplasm-localized bovine SAMHD1 proteins have similar Vpx binding capacity as human and feline SAMHD1 (Fig. 6C). Therefore, these results suggested that bovine SAMHD1 may not be degraded only through the CRL4DCAF1 E3 ubiquitin ligase complex, which is different from human and feline SAMHD1 (Fig. 7). Vpx may recruit another E3 ubiquitin ligase complex for degradation of cytoplasmic bovine SAMHD1 (Fig. 7C). Further investigation is needed to find out why only bovine SAMHD1 rather than human or feline SAMHD1 can become the substrate of this complex in the cytoplasm.
Taken together, our study supported that the lentiviral Vpx protein primarily targets nucleus-localized SAMHD1 by utilizing the E3 ubiquitin ligase complex which is also localized to the nucleus but may also have additional degradation mechanisms when countering sequencedivergent SAMHD1 proteins and suggested that feline SAMHD1 shares more common features with human SAMHD1, thus is more suitable for the establishment of the animal model to study SAMHD1 in vivo.
-
This research was funded by the National Natural Science Foundation of China (31270807), the Key Projects in the National Science & Technology Pillar Program in the Thirteenth Five-year Plan Period (2018ZX10731101-002-003 and 2018ZX10731101-001-020), Program for Jilin University Science and Technology Innovative Research Team (JLUSTIRT) (2017TD- 05), National Postdoctoral Program for Innovative Talents (BX20180124), China Postdoctoral Science Foundation (2018M64 1786), and Science and Technology Development Project of Jilin Province (20200901030SF).
-
CW, JXW and XHY designed the experiments. CW, LNM, JLW, KKZ, SZD, PYR, YZW and XYF carried out the experiments. CW and LNM analyzed the data and wrote the paper. BY, JXW and XHY checked and finalized the manuscript. CW and XHY acquired the funding. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: