
2021 Vol.36(5)
About the Special Topic on SARS-CoV-2 and COVID-19
Topic Editor: Zheng-Li Shi, PhD, Wuhan Institute of Virology, Chinese Academy of Sciences
In less than two years, SARS-CoV-2 has infected over two hundred million people and caused about 4.8 million deaths worldwide. With global efforts, over 6.3 billion vaccine doses have been administered, which largely reduce the mortality and control the virus spread. Meanwhile, the academia has never stopped the exploration to advance our knowledge in combatting the virus. In this issue, Virologica Sinica presents a collection of original articles that report the latest research progress on SARS-CoV-2, including studies on long COVID-19, development of systems for drugs and vaccines, new strategies on surveillance and counter-measures. The cover shows the basic structure of SARS-CoV-2 virus particles.
|
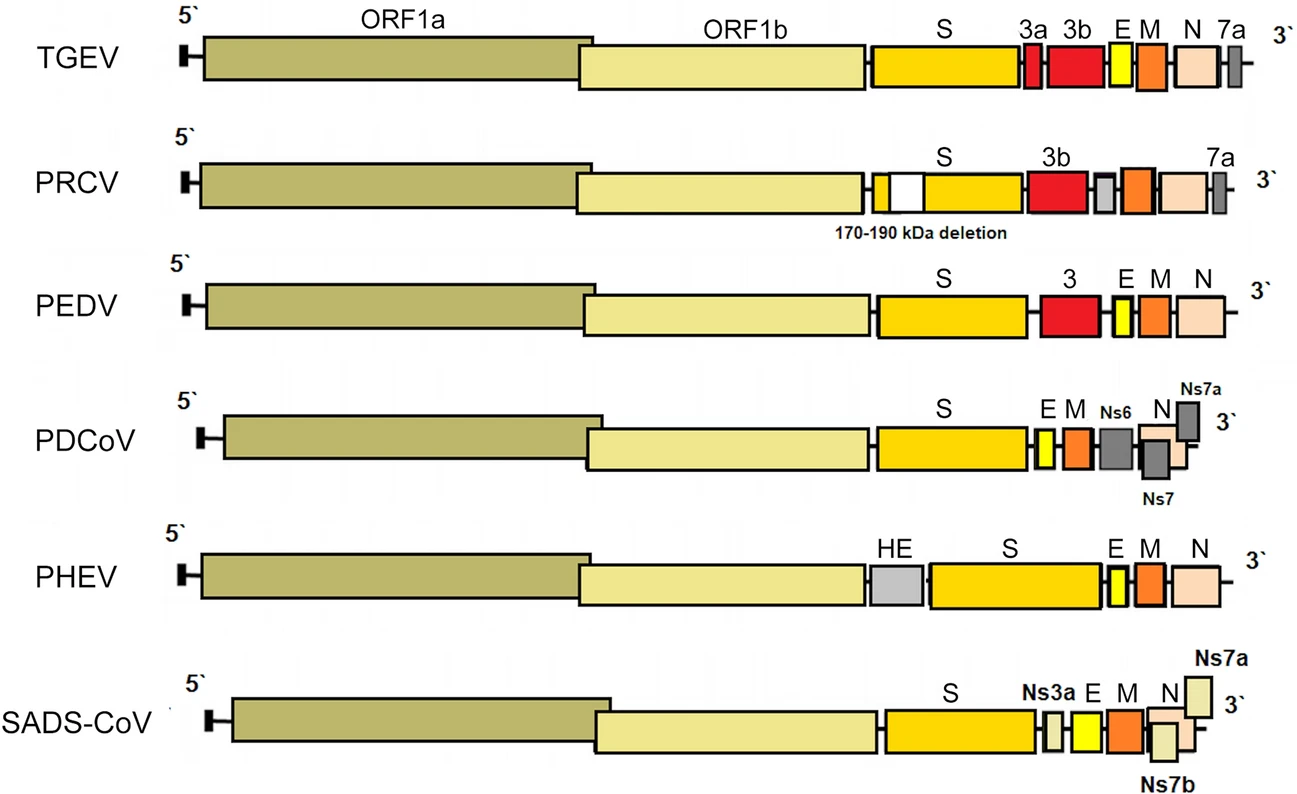
Porcine Coronaviruses: Overview of the State of the Art
2021, 36(5): 833 doi: 10.1007/s12250-021-00364-0
Like RNA viruses in general, coronaviruses (CoV) exhibit high mutation rates which, in combination with their strong tendency to recombine, enable them to overcome the host species barrier and adapt to new hosts. It is currently known that six CoV are able to infect pigs. Four of them belong to the genus Alphacoronavirus [transmissible gastroenteritis coronavirus (TEGV), porcine respiratory coronavirus (PRCV), porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronavirus (SADS-CoV)], one of them to the genus Betacoronavirus [porcine hemagglutinating encephalomyelitis virus (PHEV)] and the last one to the genus Deltacoronavirus (PDCoV). PHEV was one of the first identified swine CoV and is still widespread, causing subclinical infections in pigs in several countries. PRCV, a spike deletion mutant of TGEV associated with respiratory tract infection, appeared in the 1980s. PRCV is considered non-pathogenic since its infection course is mild or subclinical. Since its appearance, pig populations have become immune to both PRCV and TGEV, leading to a significant reduction in the clinical and economic importance of TGEV. TGEV, PEDV and PDCoV are enteropathogenic CoV and cause clinically indistinguishable acute gastroenteritis in all age groups of pigs. PDCoV and SADS-CoV have emerged in 2014 (US) and in 2017 (China), respectively. Rapid diagnosis is crucial for controlling CoV infections and preventing them from spreading. Since vaccines are available only for some porcine CoV, prevention should focus mainly on a high level of biosecurity. In view of the diversity of CoV and the potential risk factors associated with zoonotic emergence, updating the knowledge concerning this area is essential.
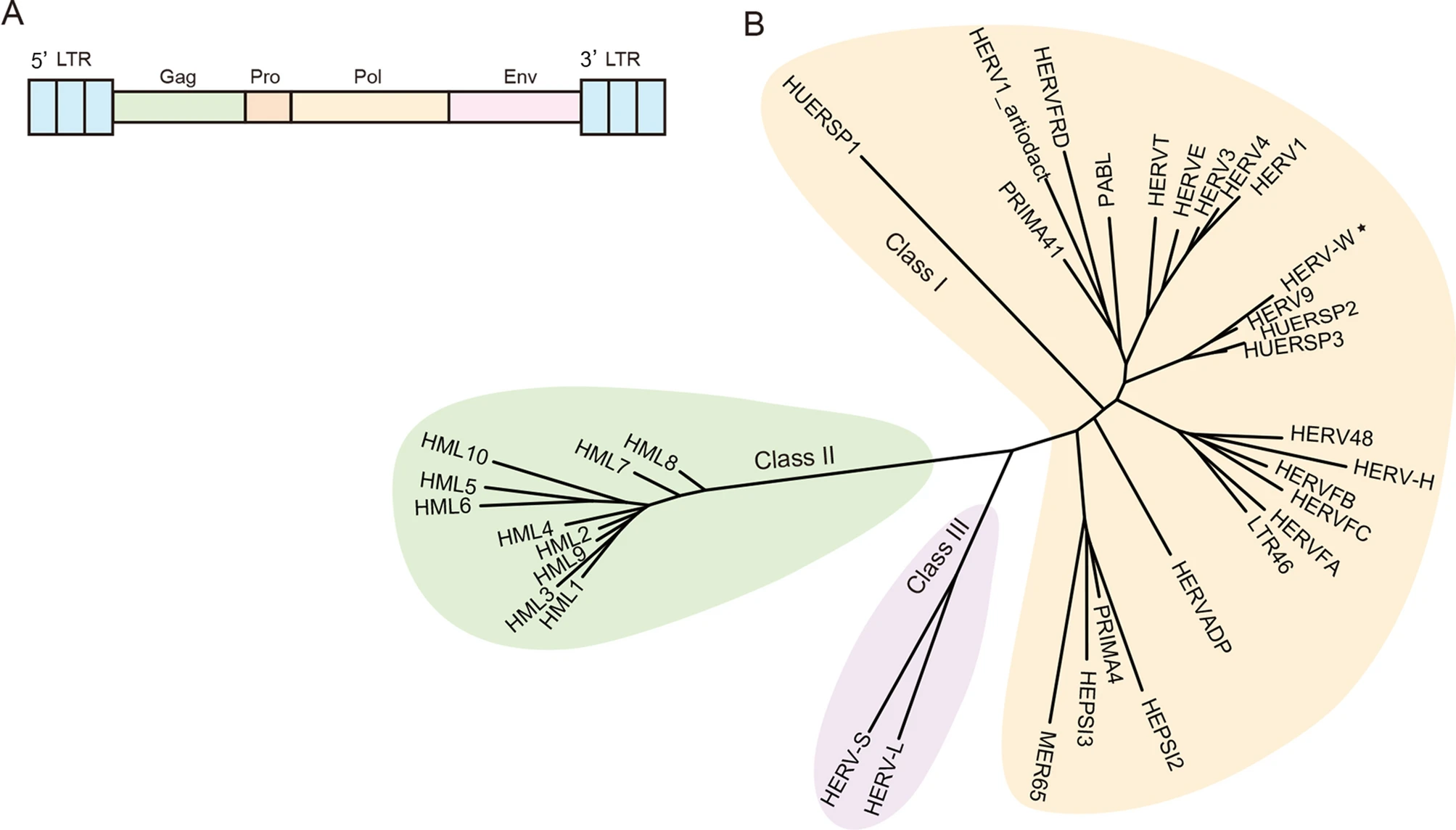
Human Endogenous Retroviruses as Biomedicine Markers
2021, 36(5): 852 doi: 10.1007/s12250-021-00387-7
Human endogenous retroviruses (HERVs) were formed via ancient integration of exogenous retroviruses into the human genome and are considered to be viral "fossils". The human genome is embedded with a considerable amount of HERVs, witnessing the long-term evolutionary history of the viruses and the host. Most HERVs have lost coding capability during selection but still function in terms of HERV-mediated regulation of host gene expression. In this review, we summarize the roles of HERV activation in response to viral infections and diseases, and emphasize the potential use of HERVs as biomedicine markers in the early diagnosis of diseases such as cancer, which provides a new perspective for the clinical application of HERVs.
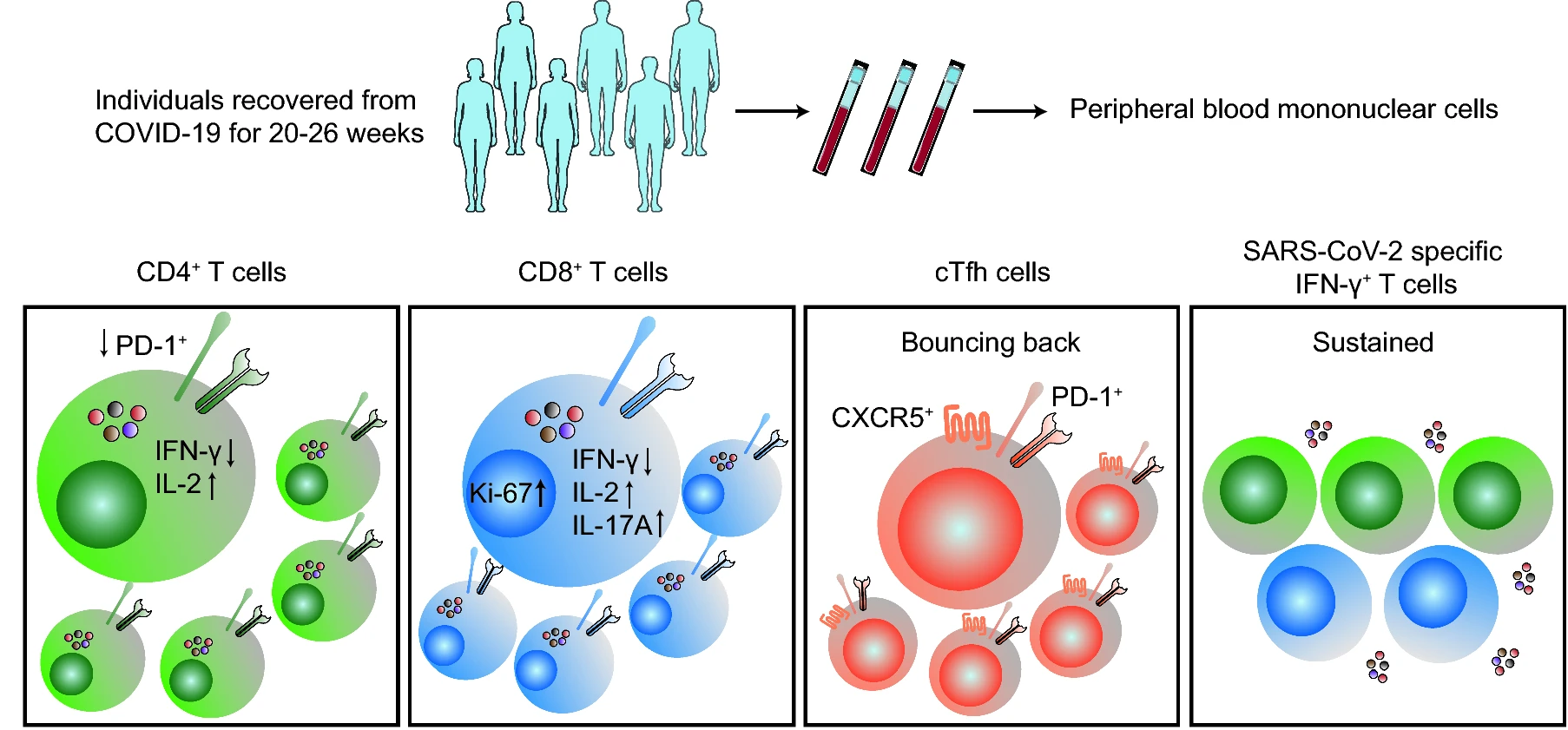
Alterations in Phenotypes and Responses of T Cells Within 6 Months of Recovery from COVID-19: A Cohort Study
2021, 36(5): 859 doi: 10.1007/s12250-021-00348-0
The COVID-19 pandemic, caused by the SARS-CoV-2 infection, is a global health crisis. While many patients have clinically recovered, little is known about long-term alterations in T cell responses of COVID-19 convalescents. In this study, T cell responses in peripheral blood mononuclear cells of a long-time COVID-19 clinically recovered (20–26 weeks) cohort (LCR) were measured via flow cytometry and ELISpot. The T cell responses of LCR were comparatively analyzed against an age and sex matched short-time clinically recovered (4–9 weeks) cohort (SCR) and a healthy donor cohort (HD). All volunteers were recruited from Wuhan Jinyintan Hospital, China. Phenotypic analysis showed that activation marker PD-1 expressing on CD4+ T cells of LCR was still significantly lower than that of HD. Functional analysis indicated that frequencies of Tc2, Th2 and Th17 in LCR were comparable to those of HD, but Tc17 was higher than that of HD. In LCR, compared to the HD, there were fewer IFN-γ producing T cells but more IL-2 secreting T cells. In addition, the circulating Tfh cells in LCR were still slightly lower compared to HD, though the subsets composition had recovered. Remarkably, SARS-CoV-2 specific T cell responses in LCR were comparable to that of SCR. Collectively, T cell responses experienced long-term alterations in phenotype and functional potential of LCR cohort. However, after clinical recovery, SARS-CoV-2 specific T cell responses could be sustained at least for six months, which may be helpful in resisting re-infection.
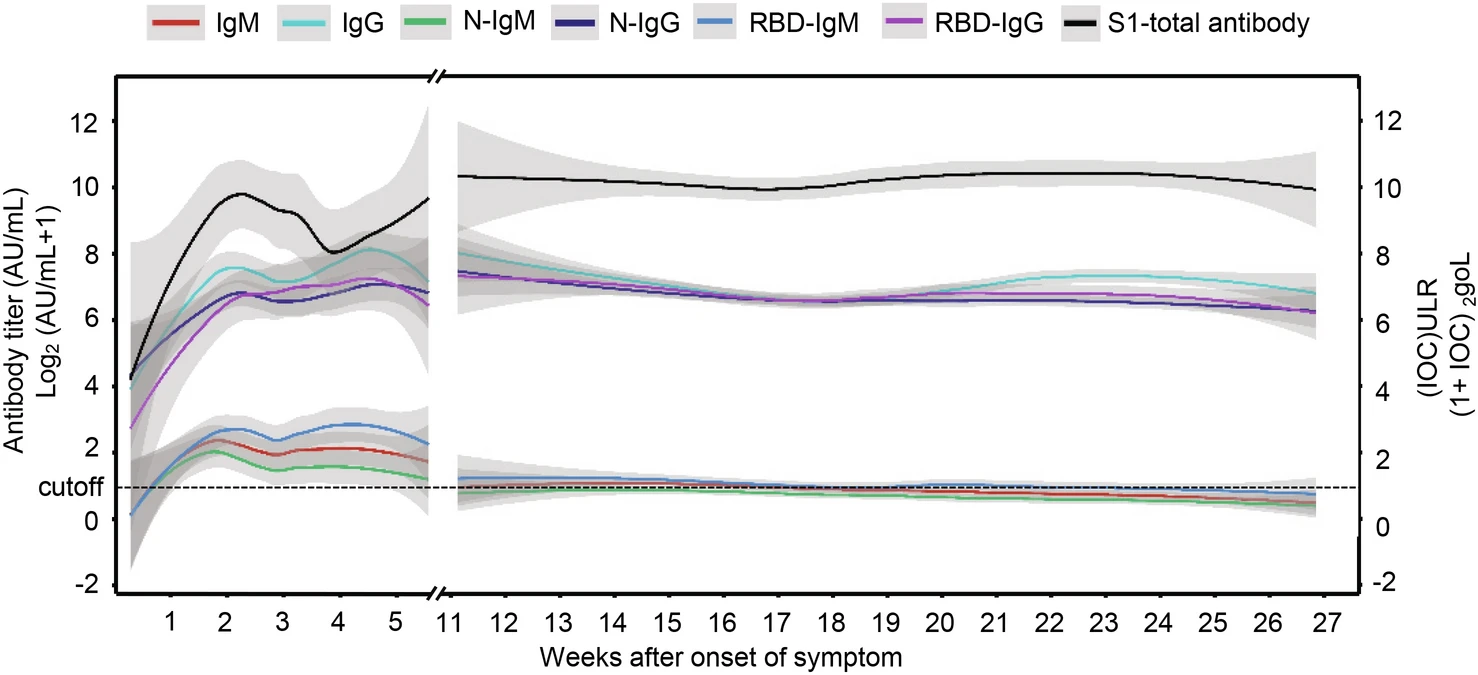
Sustainability of SARS-CoV-2 Induced Humoral Immune Responses in COVID-19 Patients from Hospitalization to Convalescence Over Six Months
2021, 36(5): 869 doi: 10.1007/s12250-021-00360-4
Understanding the persistence of antibody in convalescent COVID-19 patients may help to answer the current major concerns such as the risk of reinfection, the protection period of vaccination and the possibility of building an active herd immunity. This retrospective cohort study included 172 COVID-19 patients who were hospitalized in Wuhan. A total of 404 serum samples were obtained over six months from hospitalization to convalescence. Antibodies in the specimens were quantitatively analyzed by the capture chemiluminescence immunoassays (CLIA). All patients were positive for the anti-SARS-CoV-2 IgM/IgG at the onset of COVID-19 symptoms, and the IgG antibody persisted in all the patients during the convalescence. However, only approximately 25% of patients can detect the IgM antibodies, IgM against N protein (N-IgM) and receptor binding domain of S protein (RBD-IgM) at the 27th week. The titers of IgM, N-IgM and RBD-IgM reduced to 16.7%, 17.6% and 15.2% of their peak values respectively. In contrast, the titers of IgG, N-IgG and RBD-IgG peaked at 4–5th week and reduced to 85.9%, 62.6% and 87.2% of their peak values respectively at the end of observation. Dynamic behavior of antibodies and their correlation in age, gender and severity groups were investigated. In general, the COVID-19 antibody was sustained at high levels for over six months in most of the convalescent patients. Only a few patients with antibody reducing to an undetectable level which needs further attention. The humoral immune response against SARS-CoV-2 infection in COVID-19 patients exhibits a typical dynamic of acquired immunity.
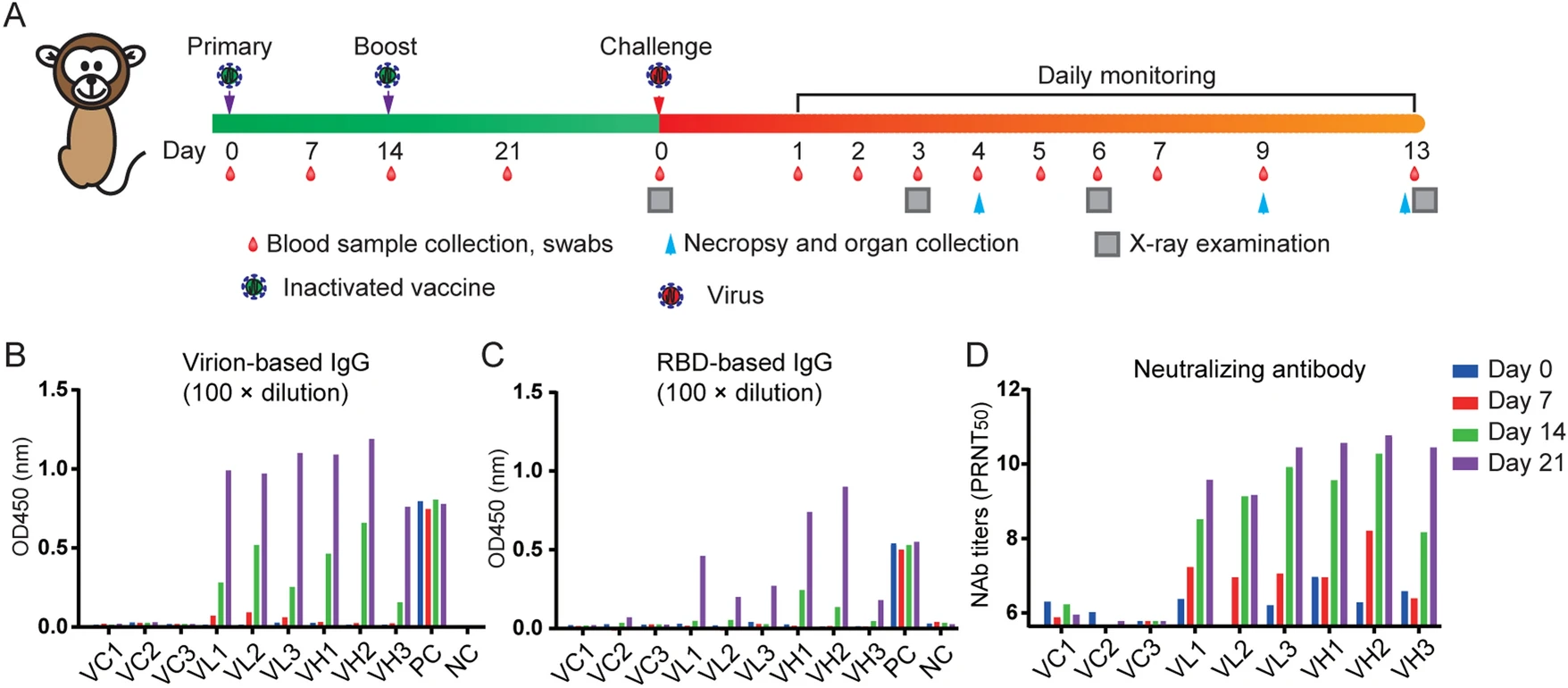
Protective Efficacy of Inactivated Vaccine against SARS-CoV-2 Infection in Mice and Non-Human Primates
2021, 36(5): 879 doi: 10.1007/s12250-021-00376-w
The ongoing coronavirus disease 2019 (COVID-19) pandemic caused more than 96 million infections and over 2 million deaths worldwide so far. However, there is no approved vaccine available for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the disease causative agent. Vaccine is the most effective approach to eradicate a pathogen. The tests of safety and efficacy in animals are pivotal for developing a vaccine and before the vaccine is applied to human populations. Here we evaluated the safety, immunogenicity, and efficacy of an inactivated vaccine based on the whole viral particles in human ACE2 transgenic mouse and in non-human primates. Our data showed that the inactivated vaccine successfully induced SARS-CoV-2-specific neutralizing antibodies in mice and non-human primates, and subsequently provided partial (in low dose) or full (in high dose) protection of challenge in the tested animals. In addition, passive serum transferred from vaccine-immunized mice could also provide full protection from SARS-CoV-2 infection in mice. These results warranted positive outcomes in future clinical trials in humans.
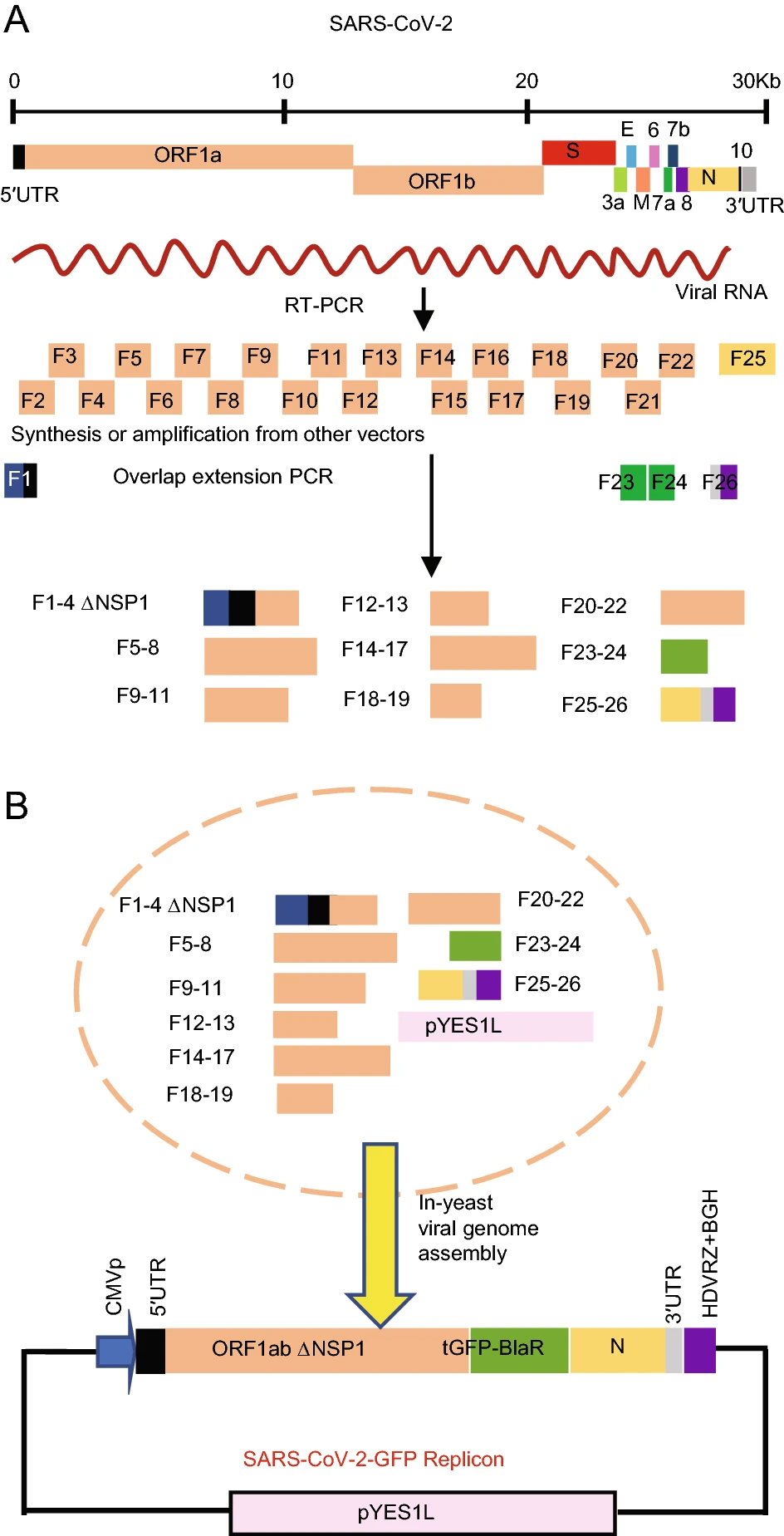
Construction of Non-infectious SARS-CoV-2 Replicons and Their Application in Drug Evaluation
2021, 36(5): 890 doi: 10.1007/s12250-021-00369-9
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a devastating pandemic worldwide. Vaccines and antiviral drugs are the most promising candidates for combating this global epidemic, and scientists all over the world have made great efforts to this end. However, manipulation of the SARS-CoV-2 should be performed in the biosafety level 3 laboratory. This makes experiments complicated and time-consuming. Therefore, a safer system for working with this virus is urgently needed. Here, we report the construction of plasmid-based, non-infectious SARS-CoV-2 replicons with turbo-green fluorescent protein and/or firefly luciferase reporters by reverse genetics using transformation-associated recombination cloning in Saccharomyces cerevisiae. Replication of these replicons was achieved simply by direct transfection of cells with the replicon plasmids as evident by the expression of reporter genes. Using SARS-CoV-2 replicons, the inhibitory effects of E64-D and remdesivir on SARS-CoV-2 replication were confirmed, and the half-maximal effective concentration (EC50) value of remdesivir and E64-D was estimated by different quantification methods respectively, indicating that these SARS-CoV-2 replicons are useful tools for antiviral drug evaluation.
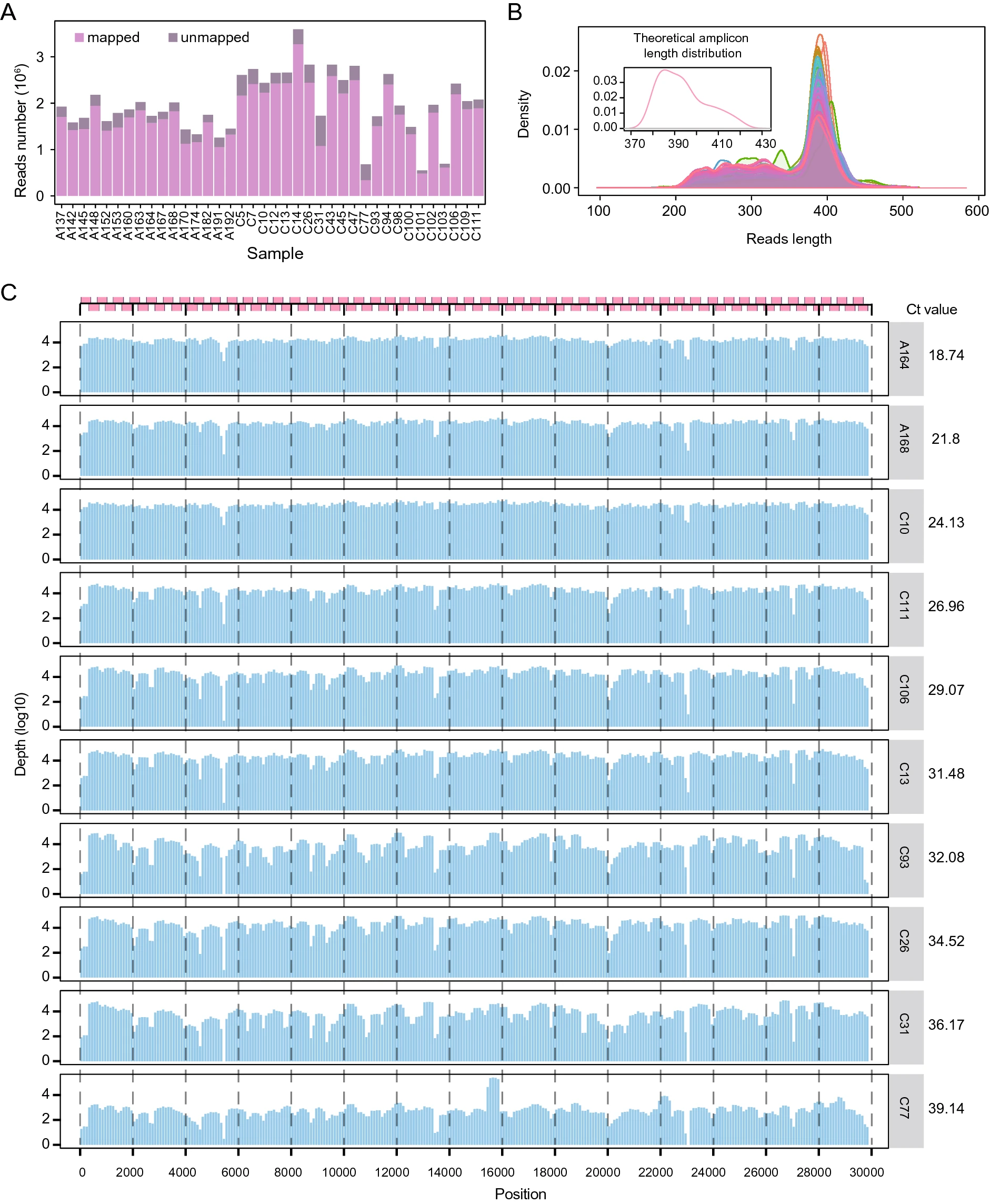
Rapid Acquisition of High-Quality SARS-CoV-2 Genome via Amplicon-Oxford Nanopore Sequencing
2021, 36(5): 901 doi: 10.1007/s12250-021-00378-8
Genome sequencing has shown strong capabilities in the initial stages of the COVID-19 pandemic such as pathogen identification and virus preliminary tracing. While the rapid acquisition of SARS-CoV-2 genome from clinical specimens is limited by their low nucleic acid load and the complexity of the nucleic acid background. To address this issue, we modified and evaluated an approach by utilizing SARS-CoV-2-specific amplicon amplification and Oxford Nanopore PromethION platform. This workflow started with the throat swab of the COVID-19 patient, combined reverse transcript PCR, and multi-amplification in one-step to shorten the experiment time, then can quickly and steadily obtain high-quality SARS-CoV-2 genome within 24 h. A comprehensive evaluation of the method was conducted in 42 samples: the sequencing quality of the method was correlated well with the viral load of the samples; high-quality SARS-CoV-2 genome could be obtained stably in the samples with Ct value up to 39.14; data yielding for different Ct values were assessed and the recommended sequencing time was 8 h for samples with Ct value of less than 20; variation analysis indicated that the method can detect the existing and emerging genomic mutations as well; Illumina sequencing verified that ultra-deep sequencing can greatly improve the single read error rate of Nanopore sequencing, making it as low as 0.4/10,000 bp. In summary, high-quality SARS-CoV-2 genome can be acquired by utilizing the amplicon amplification and it is an effective method in accelerating the acquisition of genetic resources and tracking the genome diversity of SARS-CoV-2.
A Convenient and Biosafe Replicon with Accessory Genes of SARS-CoV-2 and Its Potential Application in Antiviral Drug Discovery
2021, 36(5): 913 doi: 10.1007/s12250-021-00385-9
SARS-CoV-2 causes the pandemic of COVID-19 and no effective drugs for this disease are available thus far. Due to the high infectivity and pathogenicity of this virus, all studies on the live virus are strictly confined in the biosafety level 3 (BSL3) laboratory but this would hinder the basic research and antiviral drug development of SARS-CoV-2 because the BSL3 facility is not commonly available and the work in the containment is costly and laborious. In this study, we constructed a reverse genetics system of SARS-CoV-2 by assembling the viral cDNA in a bacterial artificial chromosome (BAC) vector with deletion of the spike (S) gene. Transfection of the cDNA into cells results in the production of an RNA replicon that keeps the capability of genome or subgenome replication but is deficient in virion assembly and infection due to the absence of S protein. Therefore, such a replicon system is not infectious and can be used in ordinary biological laboratories. We confirmed the efficient replication of the replicon by demonstrating the expression of the subgenomic RNAs which have similar profiles to the wild-type virus. By mutational analysis of nsp12 and nsp14, we showed that the RNA polymerase, exonuclease, and cap N7 methyltransferase play essential roles in genome replication and sgRNA production. We also created a SARS-CoV-2 replicon carrying a luciferase reporter gene and this system was validated by the inhibition assays with known anti-SARS-CoV-2 inhibitors. Thus, such a one-plasmid system is biosafe and convenient to use, which will benefit both fundamental research and development of antiviral drugs.
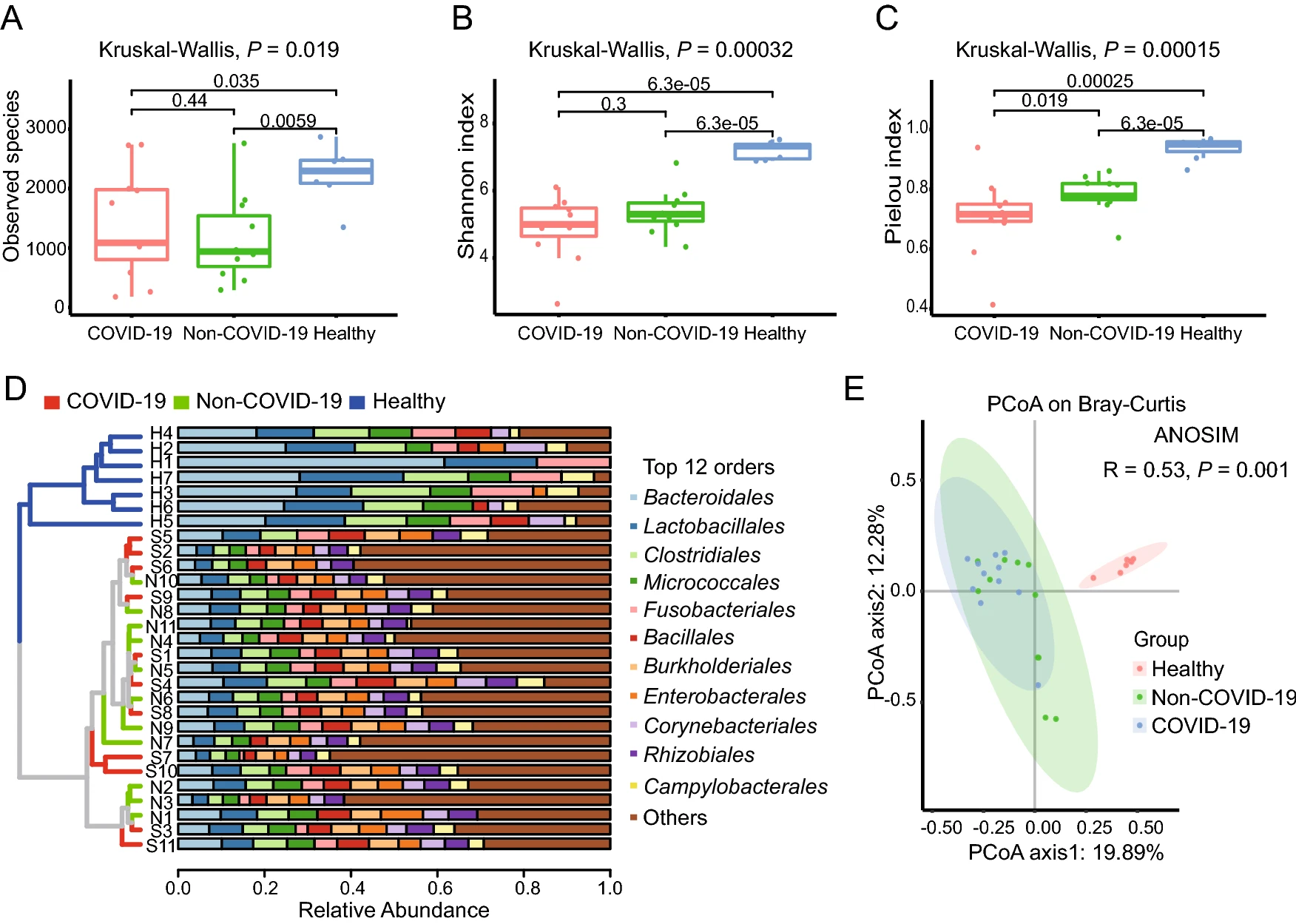
Enriched Opportunistic Pathogens Revealed by Metagenomic Sequencing Hint Potential Linkages between Pharyngeal Microbiota and COVID-19
2021, 36(5): 924 doi: 10.1007/s12250-021-00391-x
As a respiratory tract virus, SARS-CoV-2 infected people through contacting with the upper respiratory tract first. Previous studies indicated that microbiota could modulate immune response against pathogen infection. In the present study, we performed metagenomic sequencing of pharyngeal swabs from eleven patients with COVID-19 and eleven Non-COVID-19 patients who had similar symptoms such as fever and cough. Through metagenomic analysis of the above two groups and a healthy group from the public data, there are 6502 species identified in the samples. Specifically, the Pielou index indicated a lower evenness of the microbiota in the COVID-19 group than that in the Non-COVID-19 group. Combined with the linear discriminant analysis (LDA) and the generalized linear model, eighty-one bacterial species were found with increased abundance in the COVID-19 group, where 51 species were enriched more than 8 folds. The top three enriched genera were Streptococcus, Prevotella and Campylobacter containing some opportunistic pathogens. More interestingly, through experiments, we found that two Streptococcus strains, S. suis and S. agalactiae, could stimulate the expression of ACE2 of Vero cells in vitro, which may promote SARS-CoV-2 infection. Therefore, these enriched pathogens in the pharynxes of COVID-19 patients may involve in the virus-host interactions to affect SARS-CoV-2 infection and cause potential secondary bacterial infections through changing the expression of the viral receptor ACE2 and/or modulate the host's immune system.
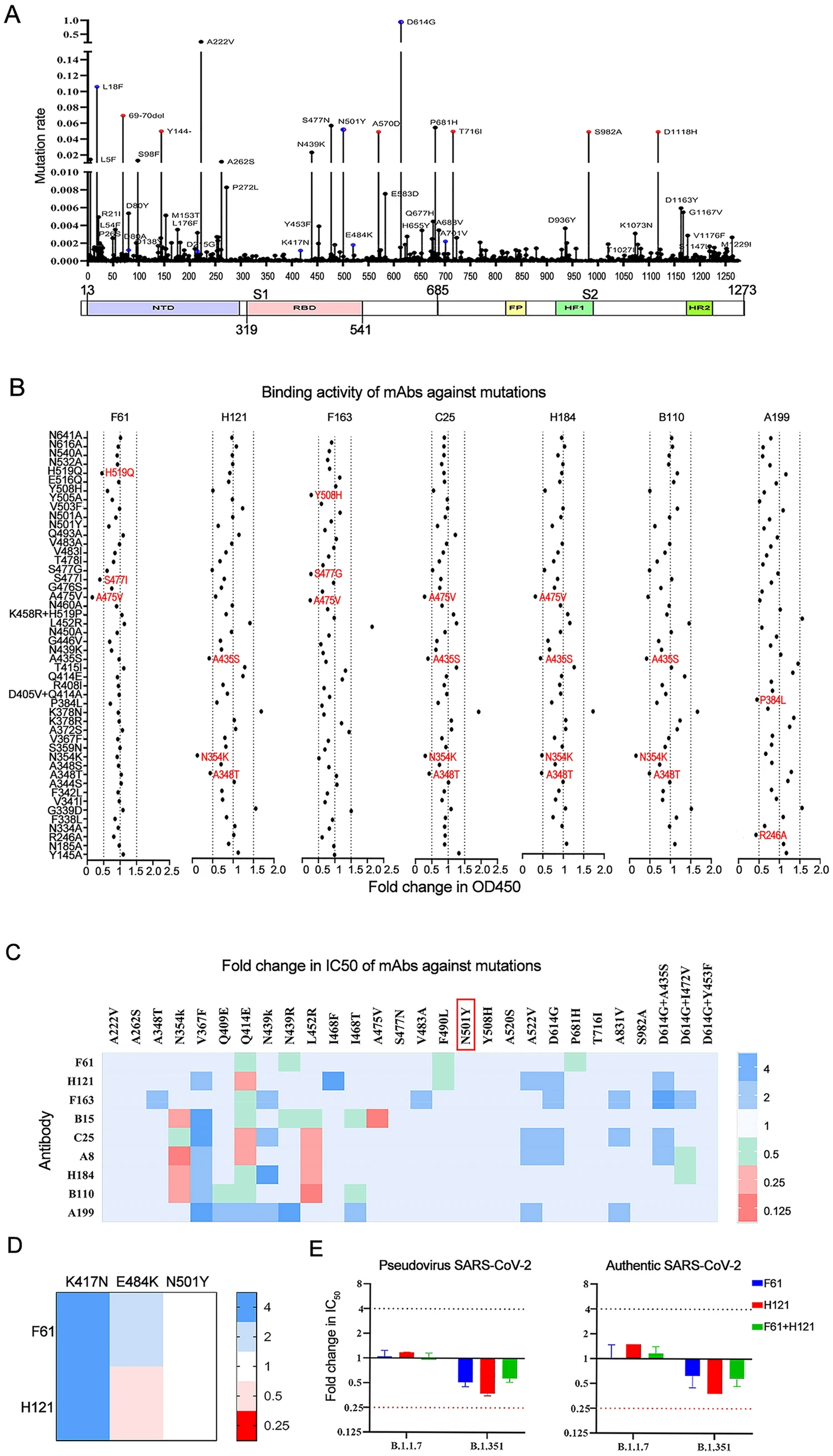
Antibody Cocktail Exhibits Broad Neutralization Activity Against SARS-CoV-2 and SARS-CoV-2 Variants
2021, 36(5): 934 doi: 10.1007/s12250-021-00409-4
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has precipitated multiple variants resistant to therapeutic antibodies. In this study, 12 high-affinity antibodies were generated from convalescent donors in early outbreaks using immune antibody phage display libraries. Of them, two RBD-binding antibodies (F61 and H121) showed high-affinity neutralization against SARS-CoV-2, whereas three S2-target antibodies failed to neutralize SARS-CoV-2. Following structure analysis, F61 identified a linear epitope located in residues G446–S494, which overlapped with angiotensin-converting enzyme 2 (ACE2) binding sites, while H121 recognized a conformational epitope located on the side face of RBD, outside from ACE2 binding domain. Hence the cocktail of the two antibodies achieved better performance of neutralization to SARS-CoV-2. Importantly, these two antibodies also showed efficient neutralizing activities to the variants including B.1.1.7 and B.1.351, and reacted with mutations of N501Y, E484K, and L452R, indicated that it may also neutralize the recent India endemic strain B.1.617. The unchanged binding activity of F61 and H121 to RBD with multiple mutations revealed a broad neutralizing activity against variants, which mitigated the risk of viral escape. Our findings revealed the therapeutic basis of cocktail antibodies against constantly emerging SARS-CoV-2 variants and provided promising candidate antibodies to clinical treatment of COVID-19 patients infected with broad SARS-CoV-2 variants.
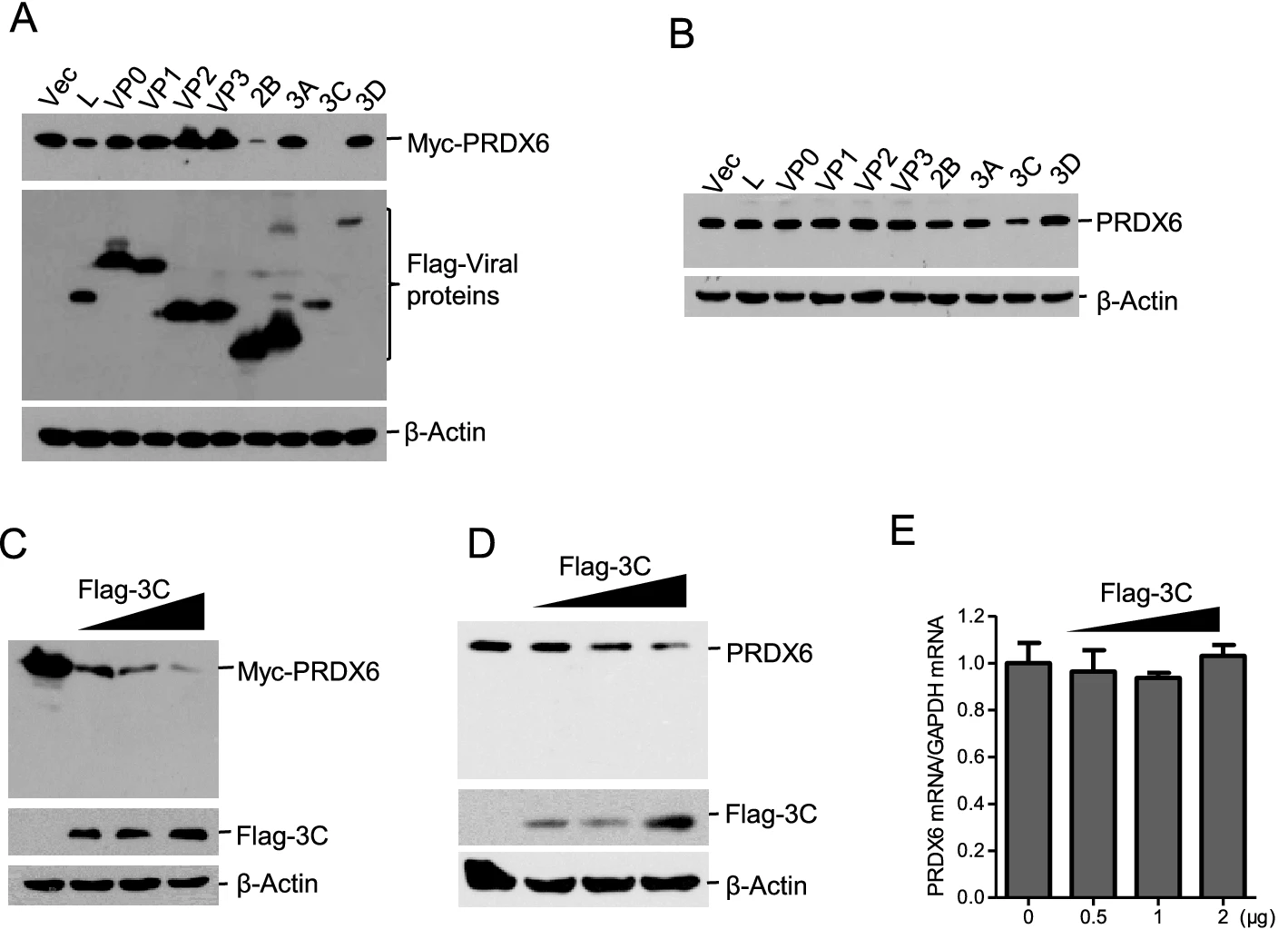
Porcine Picornavirus 3C Protease Degrades PRDX6 to Impair PRDX6-mediated Antiviral Function
2021, 36(5): 948 doi: 10.1007/s12250-021-00352-4
Peroxiredoxin-6 (PRDX6) is an antioxidant enzyme with both the activities of peroxidase and phospholipase A2 (PLA2), which is involved in regulation of many cellular reactions. However, the function of PRDX6 during virus infection remains unknown. In this study, we found that the abundance of PRDX6 protein was dramatically decreased in foot-and-mouth disease virus (FMDV) infected cells. Overexpression of PRDX6 inhibited FMDV replication. In contrast, knockdown of PRDX6 expression promoted FMDV replication, suggesting an antiviral role of PRDX6. To explore whether the activity of peroxidase and PLA2 was associated with PRDX6-mediated antiviral function, a specific inhibitor of PLA2 (MJ33) and a specific inhibitor of peroxidase activity (mercaptosuccinate) were used to treat the cells before FMDV infection. The results showed that incubation of MJ33 but not mercaptosuccinate promoted FMDV replication. Meanwhile, overexpression of PRDX6 slightly enhanced type I interferon signaling. We further determined that the viral 3Cpro was responsible for degradation of PRDX6, and 3Cpro-induced reduction of PRDX6 was independent of the proteasome, lysosome, and caspase pathways. The protease activity of 3Cpro was required for induction of PRDX6 reduction. Besides, PRDX6 suppressed the replication of another porcine picornavirus Senecavirus A (SVA), and the 3Cpro of SVA induced the reduction of PRDX6 through its proteolytic activity as well. Together, our results suggested that PRDX6 plays an important antiviral role during porcine picornavirus infection, and the viral 3Cpro induces the degradation of PRDX6 to overcome PRDX6-mediated antiviral function.
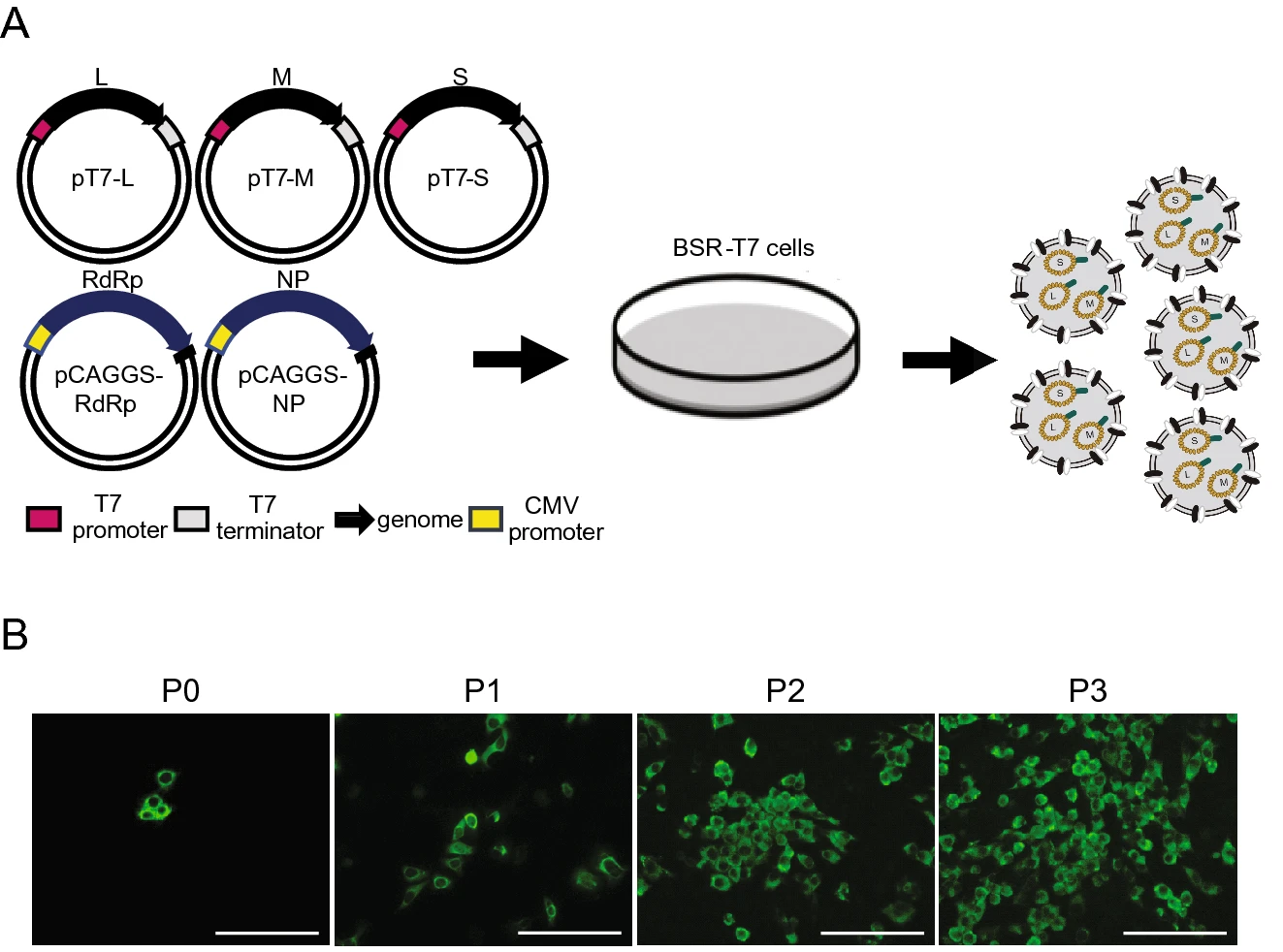
Establishment of a Reverse Genetic System of Severe Fever with Thrombocytopenia Syndrome Virus Based on a C4 Strain
2021, 36(5): 958 doi: 10.1007/s12250-021-00359-x
Severe fever with thrombocytopenia syndrome virus (SFTSV) is an emerging tick-borne bunyavirus that causes hemorrhagic fever-like disease (SFTS) in humans with a case fatality rate up to 30%. To date, the molecular biology involved in SFTSV infection remains obscure. There are seven major genotypes of SFTSV (C1-C4 and J1-J3) and previously a reverse genetic system was established on a C3 strain of SFTSV. Here, we reported successfully establishment of a reverse genetics system based on a SFTSV C4 strain. First, we obtained the 5'- and 3'-terminal untranslated region (UTR) sequences of the Large (L), Medium (M) and Small (S) segments of a laboratory-adapted SFTSV C4 strain through rapid amplification of cDNA ends analysis, and developed functional T7 polymerase-based L-, M- and S-segment minigenome assays. Then, full- length cDNA clones were constructed and infectious SFTSV were recovered from co-transfected cells. Viral infectivity, growth kinetics, and viral protein expression profile of the rescued virus were compared with the laboratory-adapted virus. Focus formation assay showed that the size and morphology of the foci formed by the rescued SFTSV were indistinguishable with the laboratory-adapted virus. However, one-step growth curve and nucleoprotein expression analyses revealed the rescued virus replicated less efficiently than the laboratory-adapted virus. Sequence analysis indicated that the difference may be due to the mutations in the laboratory-adapted strain which are more prone to cell culture. The results help us to understand the molecular biology of SFTSV, and provide a useful tool for developing vaccines and antivirals against SFTS.
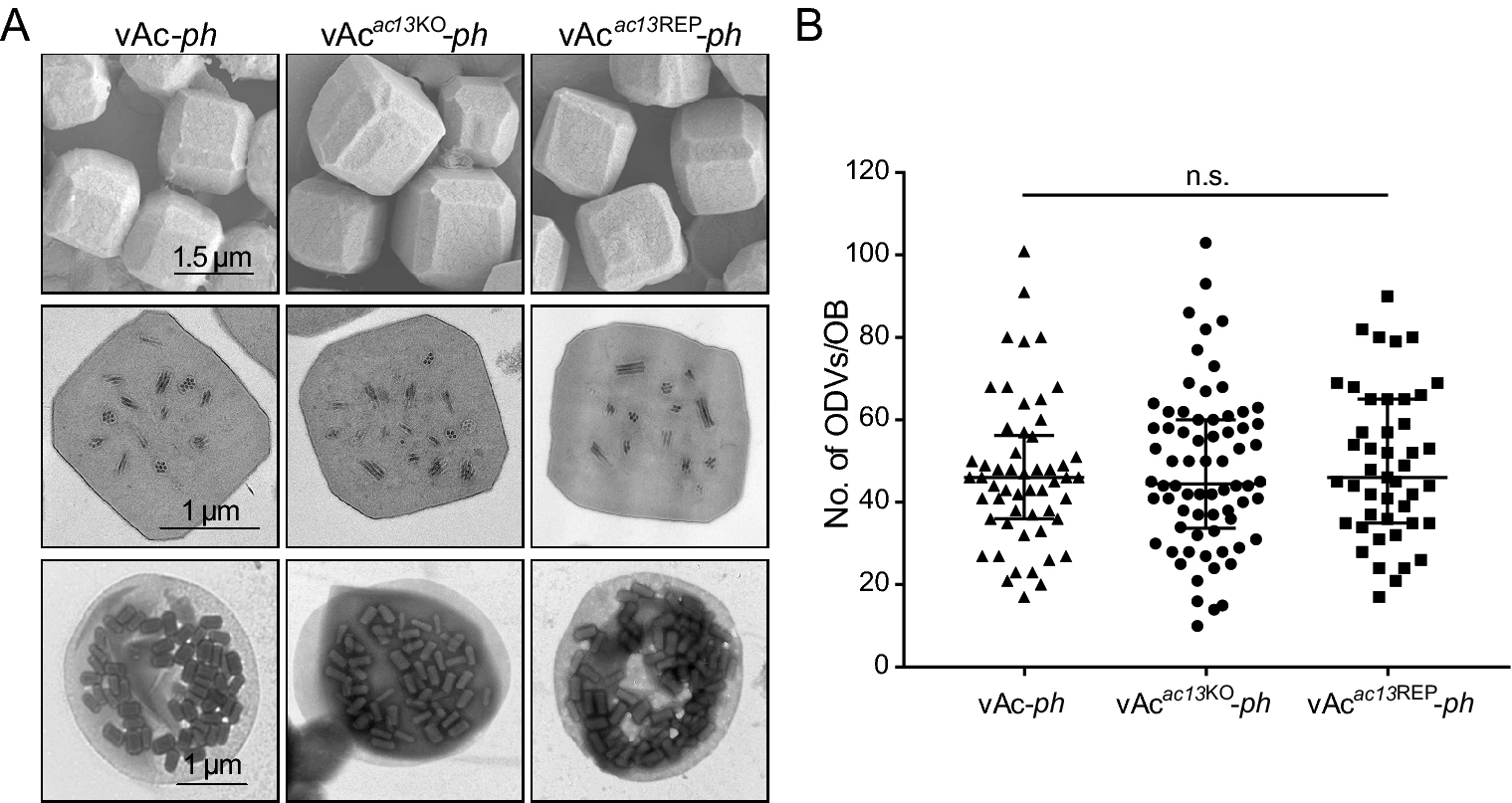
Autographa Californica Multiple Nucleopolyhedrovirus orf13 Is Required for Efficient Nuclear Egress of Nucleocapsids
2021, 36(5): 968 doi: 10.1007/s12250-021-00353-3
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) orf13 (ac13) is a conserved gene in all sequenced alphabaculoviruses. However, its function in the viral life cycle remains unknown. In this study, we found that ac13 was a late gene and that the encoded protein, bearing a putative nuclear localization signal motif, colocalized with the nuclear lamina. Deletion of ac13 did not affect viral genome replication, nucleocapsid assembly or occlusion body (OB) formation, but reduced virion budding from infected cells by approximately 400-fold compared with the wild-type virus. Deletion of ac13 substantially impaired the egress of nucleocapsids from the nucleus to the cytoplasm, while the OB morphogenesis was unaffected. Taken together, our results indicated that ac13 was required for efficient nuclear egress of nucleocapsids during virion budding, but was dispensable for OB formation.
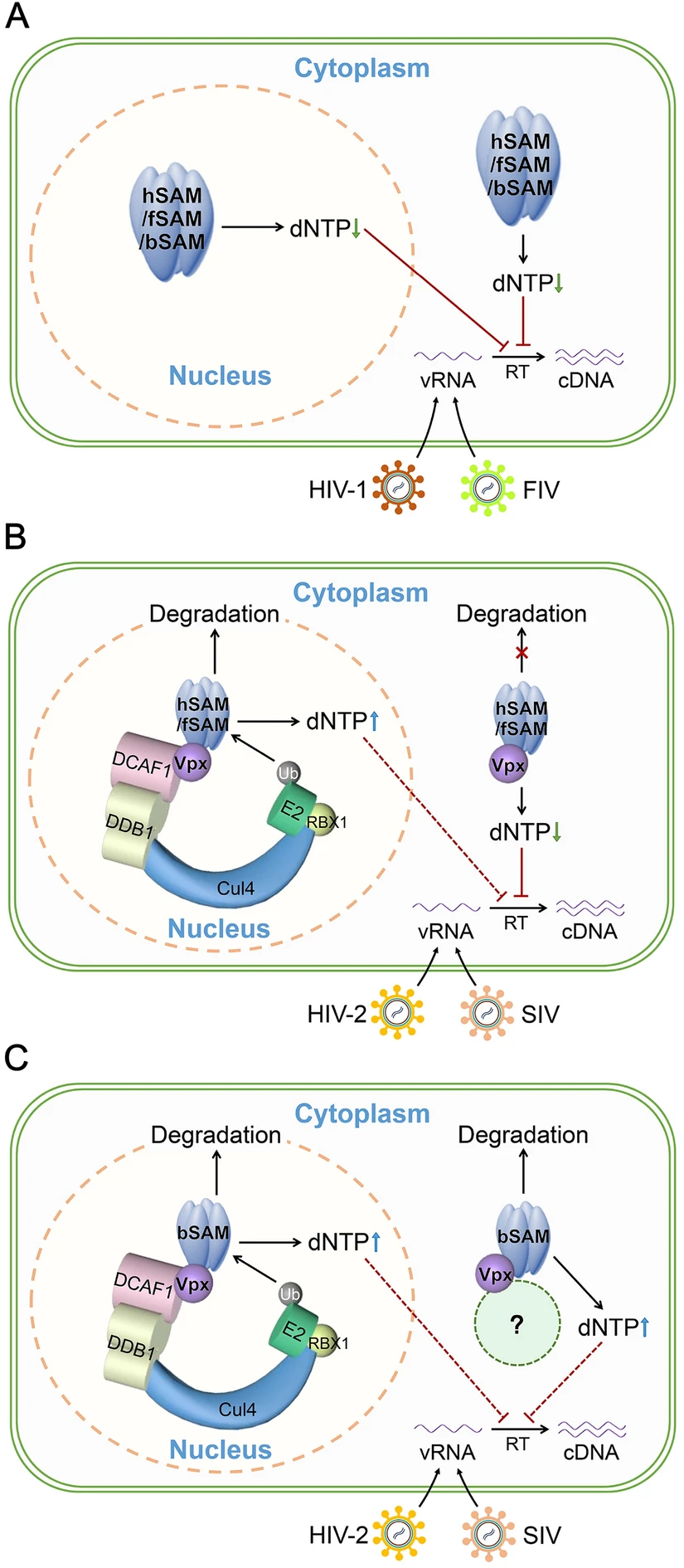
Role of Intracellular Distribution of Feline and Bovine SAMHD1 Proteins in Lentiviral Restriction
2021, 36(5): 981 doi: 10.1007/s12250-021-00351-5
Human SAMHD1 (hSAM) restricts lentiviruses at the reverse transcription step through its dNTP triphosphohydrolase (dNTPase) activity. Besides humans, several mammalian species such as cats and cows that carry their own lentiviruses also express SAMHD1. However, the intracellular distribution of feline and bovine SAMHD1 (fSAM and bSAM) and its significance in their lentiviral restriction function is not known. Here, we demonstrated that fSAM and bSAM were both predominantly localized to the nucleus and nuclear localization signal (11KRPR14)-deleted fSAM and bSAM relocalized to the cytoplasm. Both cytoplasmic fSAM and bSAM retained the antiviral function against different lentiviruses and cytoplasmic fSAM could restrict Vpx-encoding SIV and HIV-2 more efficiently than its wild-type (WT) protein as cytoplasmic hSAM. Further investigation revealed that cytoplasmic fSAM was resistant to Vpx-induced degradation like cytoplasmic hSAM, while cytoplasmic bSAM was not, but they all demonstrated the same in vitro dNTPase activity and all could interact with Vpx as their WT proteins, indicating that cytoplasmic hSAM and fSAM can suppress more SIV and HIV-2 by being less sensitive to Vpx-mediated degradation. Our results suggested that fSAM- and bSAM-mediated lentiviral restriction does not require their nuclear localization and that fSAM shares more common features with hSAM. These findings may provide insights for the establishment of alternative animal models to study SAMHD1 in vivo.
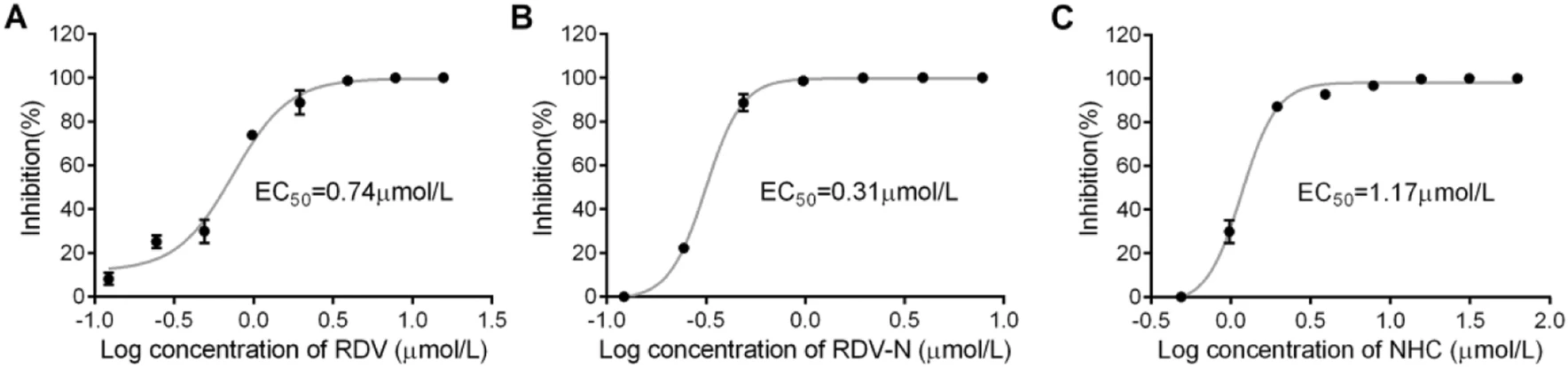
Significant Inhibition of Porcine Epidemic Diarrhea Virus In Vitro by Remdesivir, Its Parent Nucleoside and β-D-N4-hydroxycytidine
2021, 36(5): 997 doi: 10.1007/s12250-021-00362-2
Porcine epidemic diarrhea (PED) caused by porcine epidemic diarrhea virus (PEDV) is widespread in the world. In recent years, the increased virulence of the virus due to viral variations, has caused great economic losses to the pig industry in many countries. It is always worthy to find effective therapeutic methods for PED. As an important class of antivirals, nucleoside drugs which target viral polymerases have been applied in treating human viral infections for half a century. Herein, we evaluated the anti-PEDV potential of three broad-spectrum antiviral nucleoside analogs, remdesivir (RDV), its parent nucleoside (RDV-N) and β-D-N4-hydroxycytidine (NHC). Among them, RDV-N was the most active agent in Vero E6 cells with EC50 of 0.31 μmol/L, and more potent than RDV (EC50 = 0.74 μmol/L) and NHC (EC50 = 1.17 μmol/L). The activity of RDV-N was further confirmed using an indirect immuno-fluorescence assay. Moreover, RDV-N exhibited a good safety profile in cells and in mice. The high sequence similarity of the polymerase functional domains of PEDV with other five porcine coronaviruses indicated a broader antiviral spectrum for the three compounds. Generally, RDV-N is a promising broad-spectrum antiviral nucleoside, and it would be worthy to make some structural modifications to increase its oral bioavailability.
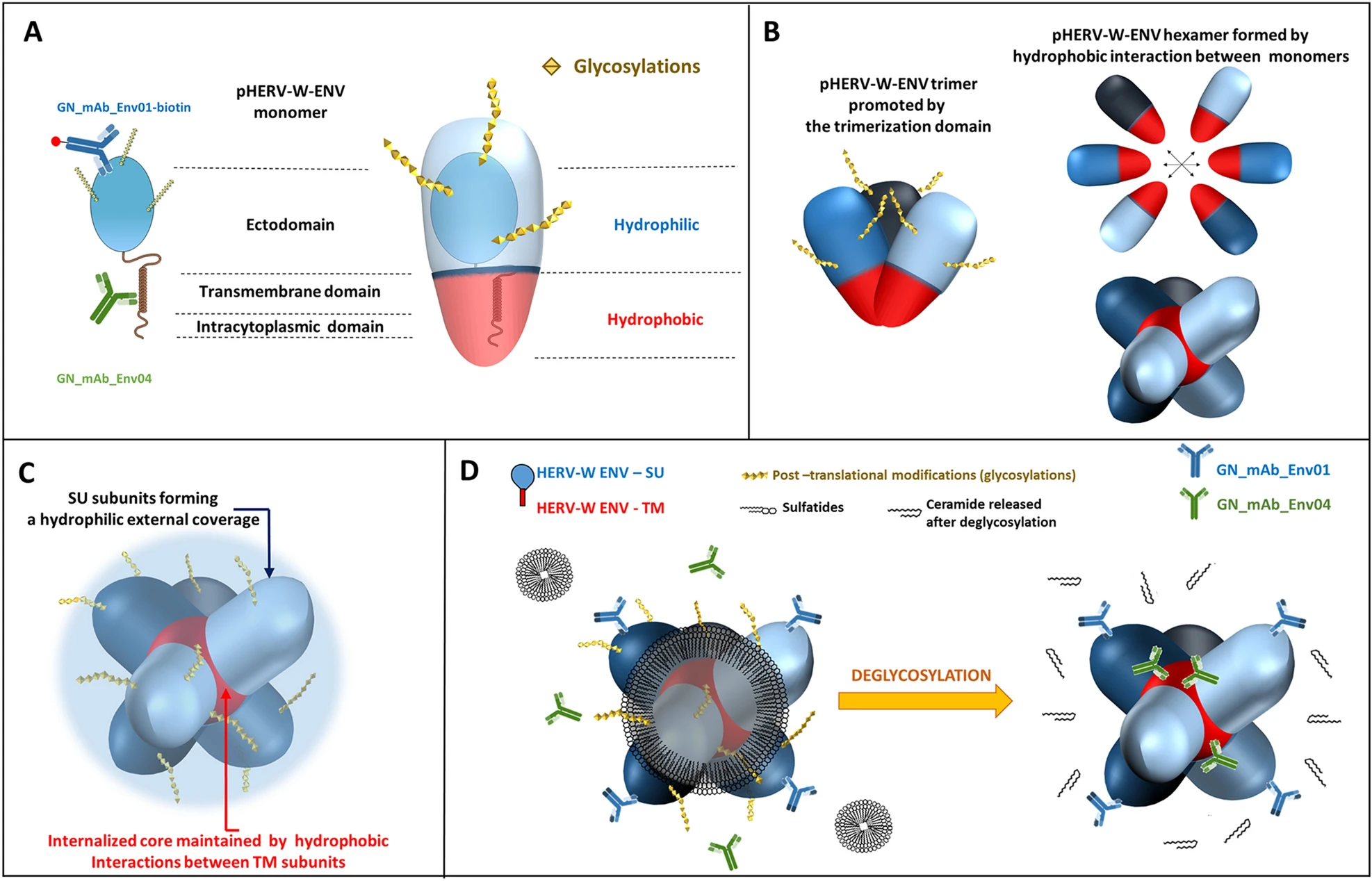
Human Endogenous Retrovirus Type W Envelope from Multiple Sclerosis Demyelinating Lesions Shows Unique Solubility and Antigenic Characteristics
2021, 36(5): 1006 doi: 10.1007/s12250-021-00372-0
In multiple sclerosis (MS), human endogenous retrovirus W family (HERV-W) envelope protein, pHERV-W ENV, limits remyelination and induces microglia-mediated neurodegeneration. To better understand its role, we examined the soluble pHERV-W antigen from MS brain lesions detected by specific antibodies. Physico-chemical and antigenic characteristics confirmed differences between pHERV-W ENV and syncytin-1. pHERV-W ENV monomers and trimers remained associated with membranes, while hexamers self-assembled from monomers into a soluble macrostructure involving sulfatides in MS brain. Extracellular hexamers are stabilized by internal hydrophobic bonds and external hydrophilic moieties. HERV-W studies in MS also suggest that this diffusible antigen may correspond to a previously described high-molecular-weight neurotoxic factor secreted by MS B-cells and thus represents a major agonist in MS pathogenesis. Adapted methods are now needed to identify encoding HERV provirus(es) in affected cells DNA. The properties and origin of MS brain pHERV-W ENV soluble antigen will allow a better understanding of the role of HERVs in MS pathogenesis. The present results anyhow pave the way to an accurate detection of the different forms of pHERV-W ENV antigen with appropriate conditions that remained unseen until now.
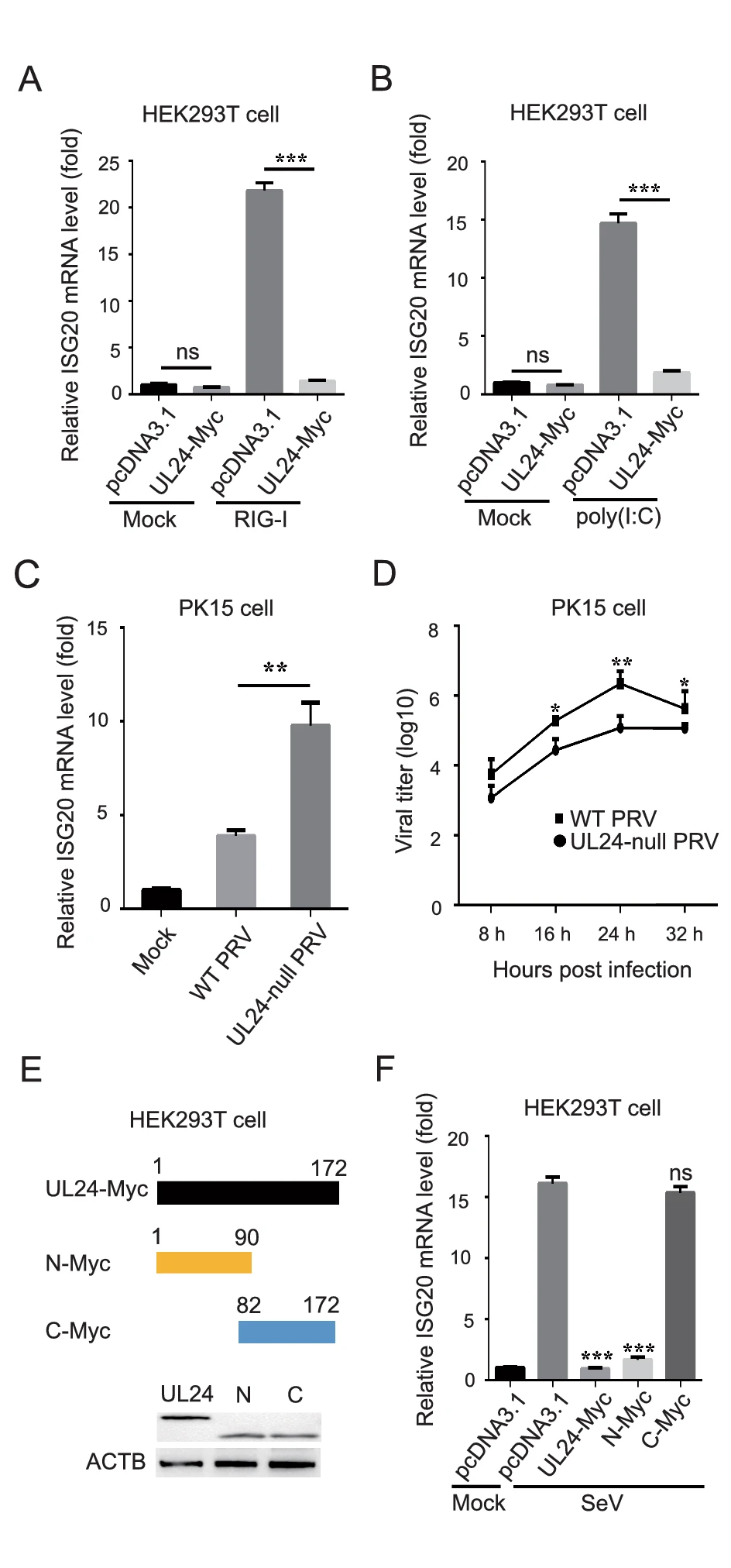
Host Interferon-Stimulated Gene 20 Inhibits Pseudorabies Virus Proliferation
2021, 36(5): 1027 doi: 10.1007/s12250-021-00380-0
Host interferon-stimulated gene 20 (ISG20) exerts antiviral effects on viruses by degrading viral RNA or by enhancing IFN signaling. Here, we examined the role of ISG20 during pseudorabies virus (PRV) proliferation. We found that ISG20 modulates PRV replication by enhancing IFN signaling. Further, ISG20 expression was upregulated following PRV infection and poly(I: C) treatment. Ectopic expression of ISG20 inhibited PRV proliferation in PK15 cells, whereas knockdown of ISG20 promoted PRV proliferation. In addition, ISG20 expression upregulated IFN-β expression and enhanced IFN downstream signaling during PRV infection. Notably, PRV UL24 suppressed the transcription of ISG20, thus antagonizing its antiviral effect. Further domain mapping analysis showed that the N terminus (amino acids 1-90) of UL24 was responsible for the inhibition of ISG20 transcription. Collectively, these findings characterize the role of ISG20 in suppressing PRV replication and increase the understanding of host-PRV interplay.
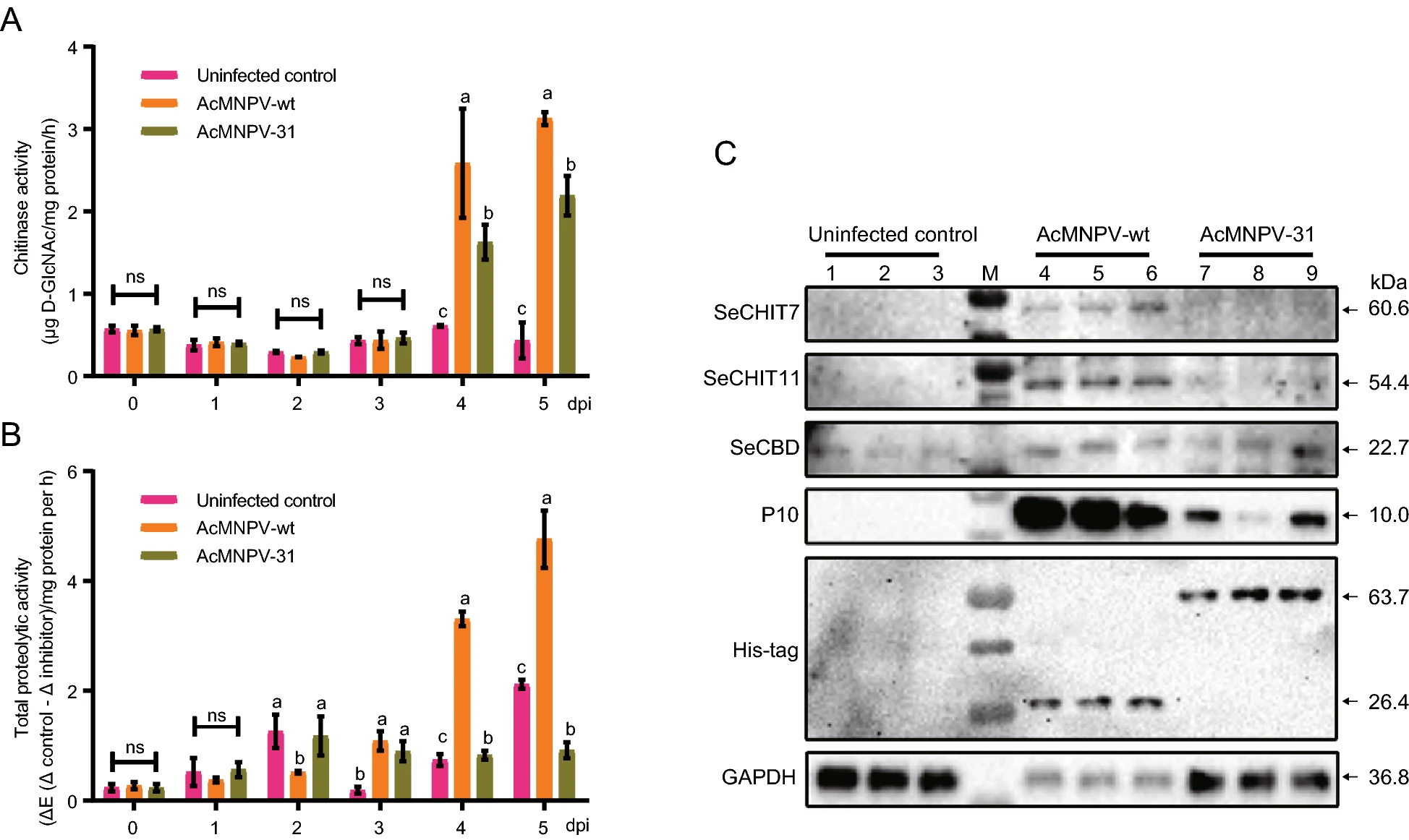
3H-31, A Non-structural Protein of Heliothis virescens ascovirus 3h, Inhibits the Host Larval Cathepsin and Chitinase Activities
2021, 36(5): 1036 doi: 10.1007/s12250-021-00374-y
3h-31 of Heliothis virescens ascovirus 3h (HvAV-3h) is a highly conserved gene of ascoviruses. As an early gene of HvAV-3h, 3h-31 codes for a non-structural protein (3H-31) of HvAV-3h. In the study, 3h-31 was initially transcribed and expressed at 3 h post-infection (hpi) in the infected Spodoptera exigua fat body cells (SeFB). 3h-31 was further inserted into the bacmid of Autographa californica nucleopolyhedrovirus (AcMNPV) to generate an infectious baculovirus (AcMNPV-31). In vivo experiments showed that budded virus production and viral DNA replication decreased with the expression of 3H-31, and lucent tubular structures were found around the virogenic stroma in the AcMNPV-31-infected SeFB cells. In vivo, both LD50 and LD90 values of AcMNPV-31 were significantly higher than those of the wild-type AcMNPV (AcMNPV-wt) in third instar S. exigua larvae. An interesting finding was that the liquefaction of the larvae killed by the infection of AcMNPV-31 was delayed. Chitinase and cathepsin activities of AcMNPV-31-infected larvae were significantly lower than those of AcMNPV-wt-infected larvae. The possible regulatory function of the chitinase and cathepsin for 3H-31 was further confirmed by RNAi, which showed that larval cathepsin activity was significantly upregulated, but chitinase activity was not significantly changed due to the RNAi of 3h-31. Based on the obtained results, we assumed that the function of 3H-31 was associated with the inhibition of host larval chitinase and cathepsin activities, so as to restrain the hosts in their larval stages.
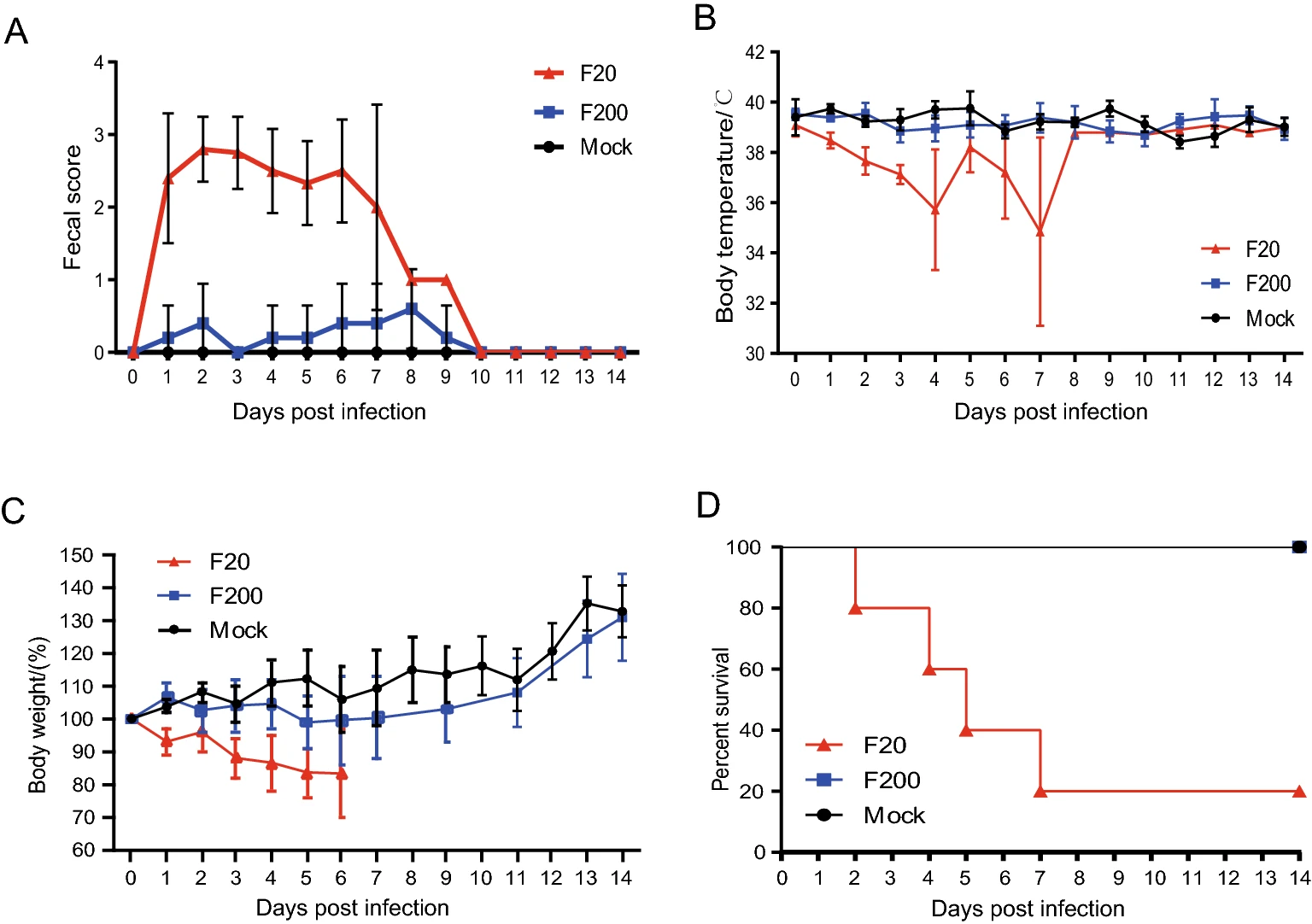
A Virulent PEDV Strain FJzz1 with Genomic Mutations and Deletions at the High Passage Level Was Attenuated in Piglets via Serial Passage In Vitro
2021, 36(5): 1052 doi: 10.1007/s12250-021-00368-w
Highly virulent porcine epidemic diarrhea virus (PEDV) strains re-emerged and circulated in China at the end of 2010, causing significant economic losses in the pork industry worldwide. To understand the genetic dynamics of PEDV during its passage in vitro, the PEDV G2 strain FJzz1 was serially propagated in Vero cells for up to 200 passages. The susceptibility and adaptability of the FJzz1 strain increased gradually as it was serially passaged in vitro. Sequence analysis revealed that amino acid (aa) changes were mainly concentrated in the S glycoprotein, which accounted for 72.22%–85.71% of all aa changes. A continuous aa deletion (55I56G57E→55K56Δ57Δ) occurred in the N-terminal domain of S1 (S1-NTD). To examine how the aa changes affected its virulence, FJzz1-F20 and FJzz1-F200 were selected to simultaneously evaluate their pathogenicity in suckling piglets. All the piglets in the FJzz1-F20-infected group showed typical diarrhea at 24 h postinfection, and the piglets died successively by 48 h postinfection. However, the clinical signs of the piglets in the FJzz1-F200-infected group were significantly weaker, and no deaths occurred. The FJzz1-F200-infected group also showed a lower level of fecal viral shedding and lower viral loads in the intestinal tissues, and no obvious histopathological lesions. Type I and III interferon were induced in the FJzz1-F200 infection group, together with pro-inflammatory cytokines, such as TNF-α, IL-1β and IL-8. These results indicate that the identified genetic changes may contribute to the attenuation of FJzz1 strain, and the attenuated FJzz1-F200 may have the potential for developing PEDV live-attenuated vaccines.
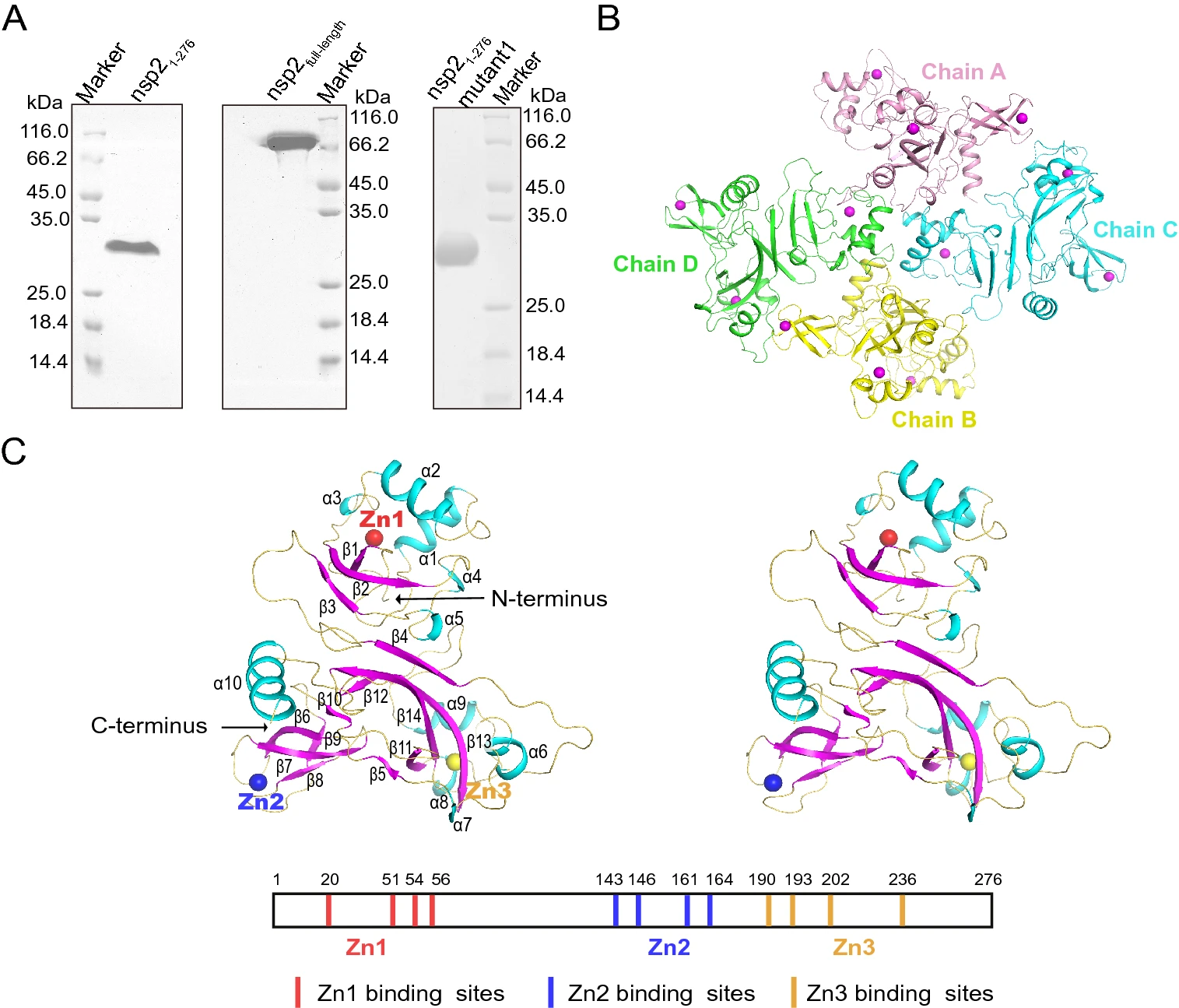
Structure and Function of N-Terminal Zinc Finger Domain of SARS-CoV-2 NSP2
2021, 36(5): 1104 doi: 10.1007/s12250-021-00431-6
SARS-CoV-2 has become a global pandemic threatening human health and safety. It is urgent to find effective therapeutic agents and targets with the continuous emergence of novel mutant strains. The knowledge of the molecular basis and pathogenesis of SARS-CoV-2 in host cells requires to be understood comprehensively. The unknown structure and function of nsp2 have hindered our understanding of its role in SARS-CoV-2 infection. Here, we report the crystal structure of the N-terminal of SARS-CoV-2 nsp2 to a high resolution of 1.96Å. This novel structure contains three zinc fingers, belonging to the C2H2, C4, and C2HC types, respectively. Structure analysis suggests that nsp2 may be involved in binding nucleic acids and regulating intracellular signaling pathways. The binding to single or double-stranded nucleic acids was mainly through the large positively charged region on the surface of nsp2, and K111, K112, K113 were key residues. Our findings lay the foundation for a better understanding of the relationship between structure and function for nsp2. It is helpful to make full use of nsp2 as further research and development of antiviral targets and drug design.
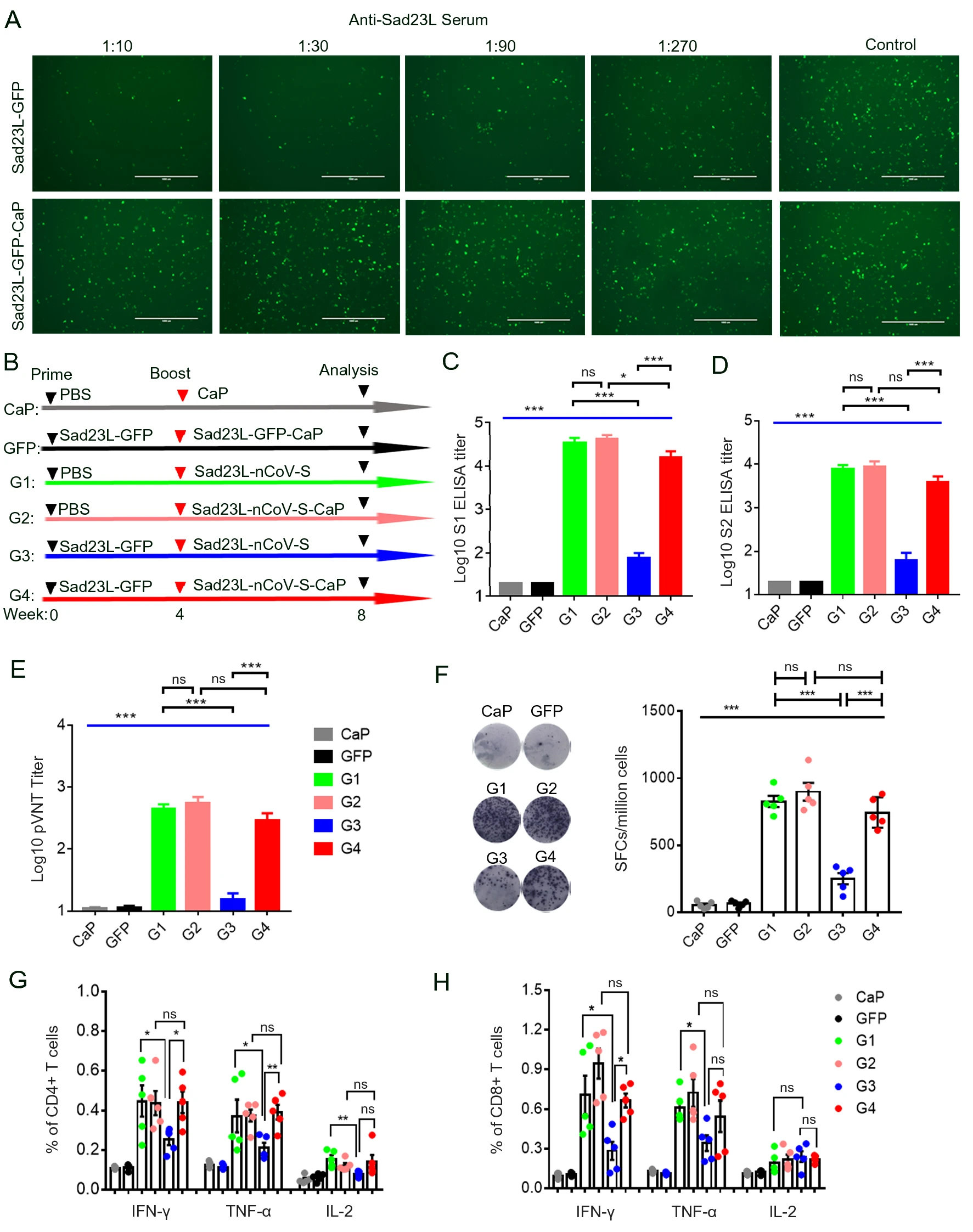
A Self-Biomineralized Novel Adenovirus Vectored COVID-19 Vaccine for Boosting Immunization of Mice
2021, 36(5): 1113 doi: 10.1007/s12250-021-00434-3
SARS-CoV-2 has caused more than 3.8 million deaths worldwide, and several types of COVID-19 vaccines are urgently approved for use, including adenovirus vectored vaccines. However, the thermal instability and pre-existing immunity have limited its wide applications. To circumvent these obstacles, we constructed a self-biomineralized adenovirus vectored COVID-19 vaccine (Sad23L-nCoV-S-CaP) by generating a calcium phosphate mineral exterior (CaP) based on Sad23L vector carrying the full-length gene of SARS-CoV-2 spike protein (S) under physiological condition. This Sad23L-nCoV-S-CaP vaccine was examined for its characteristics of structure, thermostability, immunogenicity and avoiding the problem of preexisting immunity. In thermostability test, Sad23L-nCoV-S-CaP could be stored at 4 ℃ for over 45 days, 26 ℃ for more than 8 days and 37 ℃ for approximately 2 days. Furthermore, Sad23L-nCoV-S-CaP induced higher level of S-specific antibody and T cell responses, and was not affected by the pre-existing anti-Sad23L immunity, suggesting it could be used as boosting immunization on Sad23L-nCoV-S priming vaccination. The boosting with Sad23L-nCoV-S-CaP vaccine induced high titers of 105.01 anti-S1, 104.77 anti-S2 binding antibody, 103.04 pseudovirus neutralizing antibody (IC50), and robust T-cell response of IFN-γ (1466.16 SFCs/106 cells) to S peptides, respectively. In summary, the self-biomineralization of the COVID-19 vaccine Sad23L-nCoV-S-CaP improved vaccine efficacy, which could be used in prime-boost regimen for prevention of SARS-CoV-2 infection in humans.
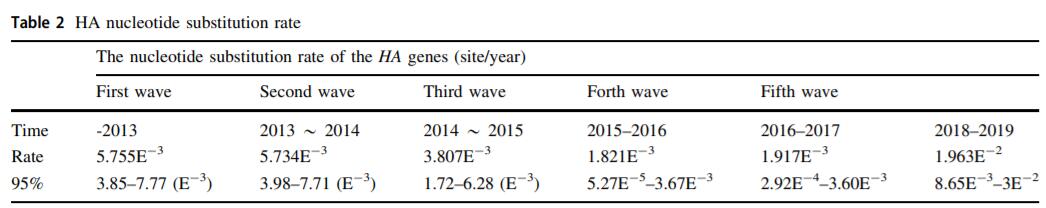
Accelerated Evolution of H7N9 Subtype Influenza Virus under Vaccination Pressure
2021, 36(5): 1124 doi: 10.1007/s12250-021-00383-x
No avian H7N9 outbreaks have occurred since the introduction of H7N9 inactivated vaccine in the fall of 2017. However, H7N9 is still prevalent in poultry. To surveil the prevalence, genetic characteristics, and antigenic changes of H7N9, over 7000 oropharyngeal and cloaca swab specimens were collected from live poultry markets and farms in 15 provinces of China from 2017 to 2019. A total of 85 influenza virus subtype H7N9 strains were isolated and 20 representative strains were selected for genetic analysis and antigenicity evaluation. Results indicated the decreased prevalence of low-pathogenic H7N9 strains while highly-pathogenic H7N9 strains became dominated since the introduction of vaccine. Phylogenetic analysis showed that strains from 2019 formed an independent small branch and were genetically distant to strains isolated in 2013–2018. Analysis of key amino acid sites showed that the virus strains may adapt to the host environment evolutionally through mutation. Our analysis predicted additional potential glycosylation sites for HA and NA genes in the 2019 strains. Sequence analysis of HA gene in strains isolated from 2018 to 2019 showed that there were an increased nucleotide substitution rate and an increased mutation rate in the first and second nucleotides of coding codons within the open reading frame. The hemagglutination inhibition (HI) assay showed that H7-Re1 and H7-Re2 exhibited a lower HI titer for isolates from 2019, while H7-Re3 and rLN79 showed a high HI titer. The protective effect of the vaccine decreased after 15 months of use. Overall, under vaccination pressure, the evolution of influenza virus subtype H7N9 has accelerated.
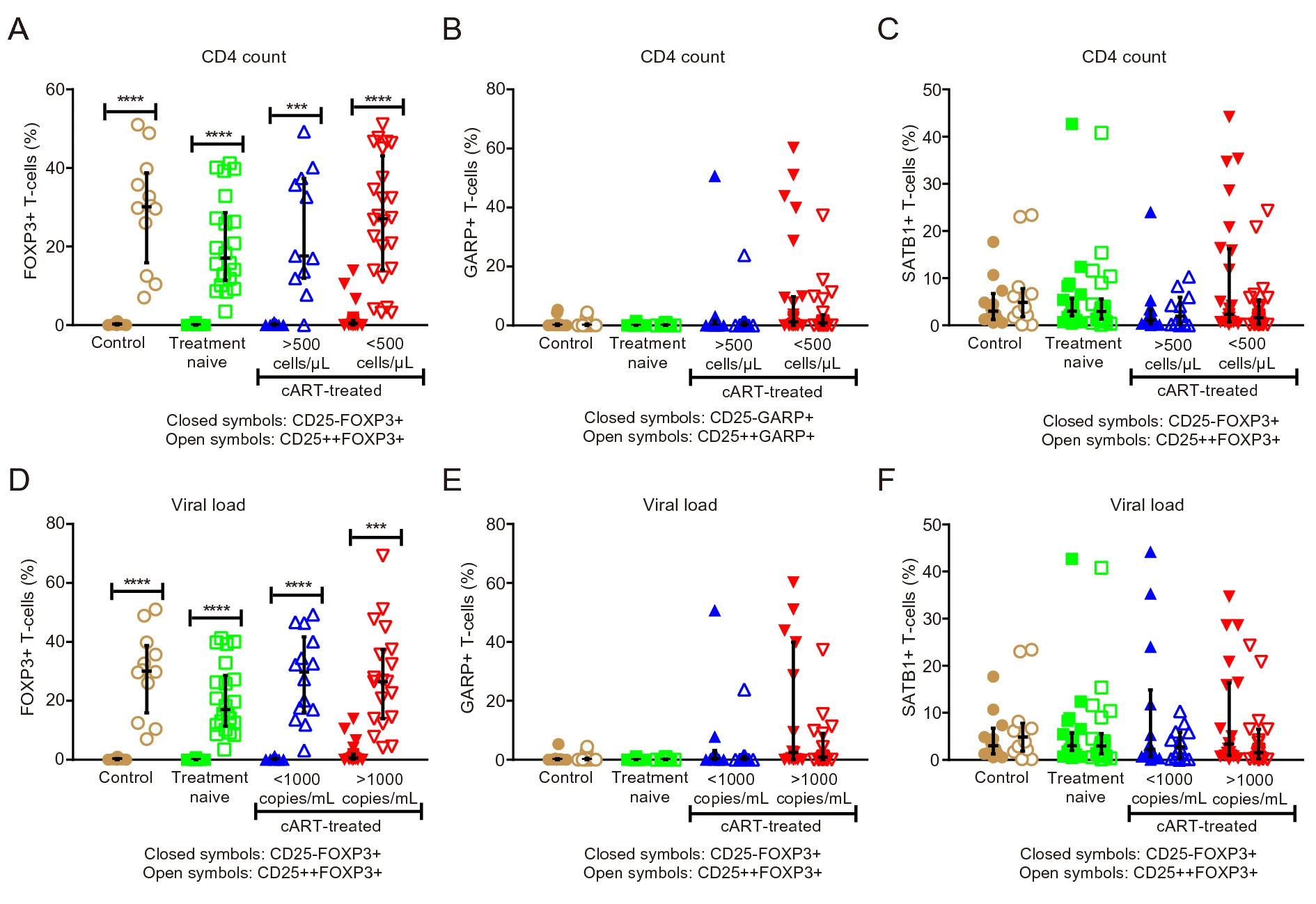
Expansion of GARP-Expressing CD4+CD25-FoxP3+ T Cells and SATB1 Association with Activation and Coagulation in Immune Compromised HIV-1-Infected Individuals in South Africa
2021, 36(5): 1133 doi: 10.1007/s12250-021-00386-8
Although antiretroviral treatment lowers the burden of human immunodeficiency virus (HIV)-related disease, it does not always result in immunological recovery. This manifests as persistent chronic inflammation, immune activation or exhaustion that can promote the onset of co-morbidities. As the exact function of regulatory T (Treg) cells in HIV remains unclear, this cross-sectional study investigated three expression markers (Forkhead box protein P3 [FOXP3], glycoprotein A repetitions predominant [GARP], special AT-rich sequence binding protein 1 [SATB1]) and compared their expansion between CD4+CD25- and CD4+CD25++ T cells. Age-matched study subjects were recruited (Western Cape, South Africa) and sub-divided: HIV-negative subjects (n = 12), HIV-positive naïve treated (n = 22), HIV-positive treated based on CD4 count cells/μL (CD4 > 500 and CD4 < 500) (n = 34) and HIV-treated based on viral load (VL) copies/mL (VL < 1000 and VL > 1000) (n = 34). Markers of immune activation (CD38) and coagulation (CD142) on T cells (CD8) were assessed by flow cytometry together with FOXP3, GARP and SATB1 expression on CD4+CD25- and CD4+CD25++ T cells. Plasma levels of interleukin-10 (IL-10; anti-inflammatory marker), IL-6 (inflammatory marker) and D-dimer (coagulation marker) were assessed. This study revealed three major findings in immuno-compromised patients with virological failure (CD4 < 500; VL > 1000): (1) the expansion of the unconventional Treg cell subset (CD4+CD25-FOXP3+) is linked with disease progression markers; (2) increased GARP expression in the CD4+CD25- and CD4+CD25++ subsets; and (3) the identification of a strong link between CD4+CD25-SATB1+ cells and markers of immune activation (CD8+CD38+) and coagulation (CD8+CD142+ and D-dimer).
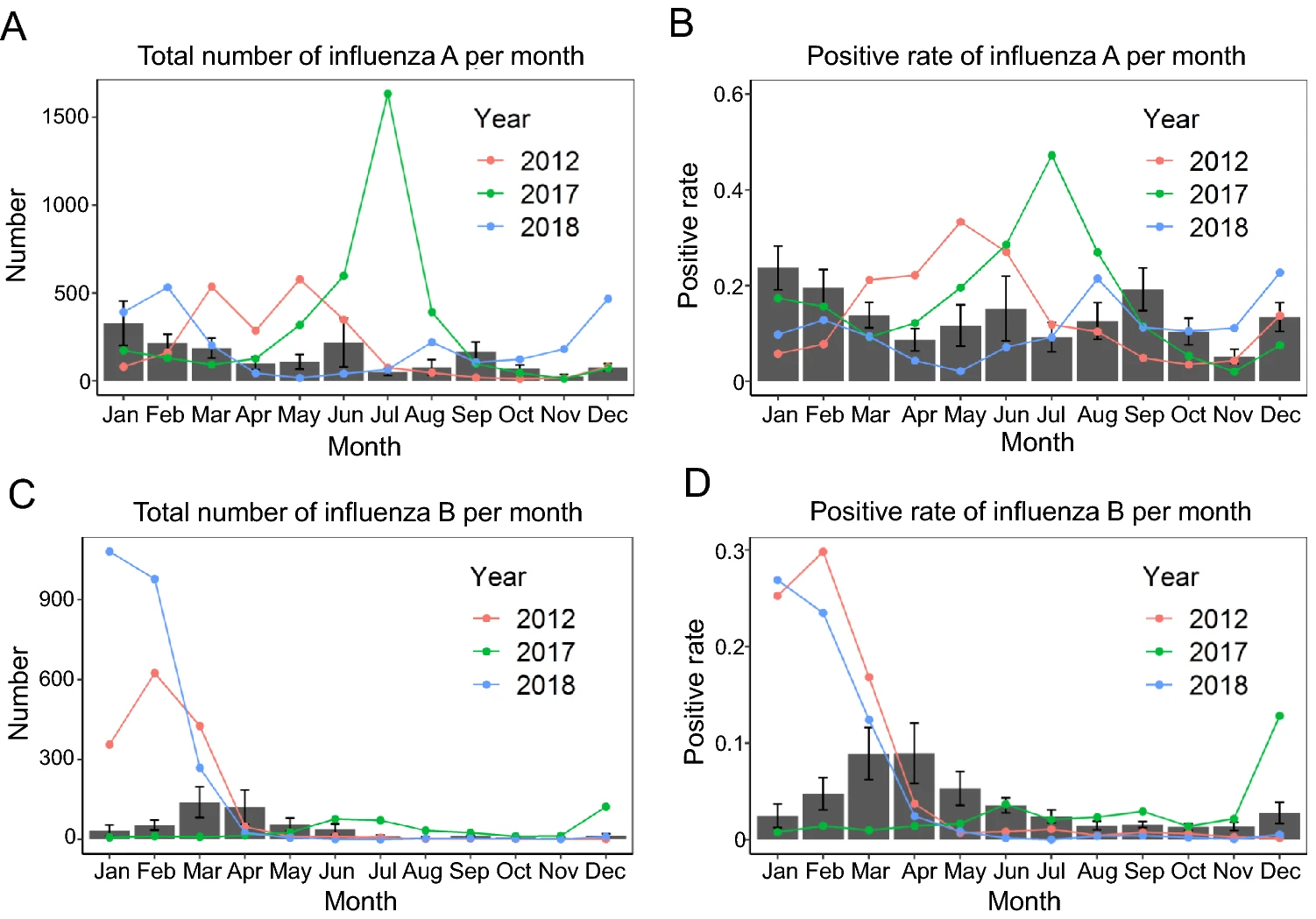
Epidemiological Characteristics of Influenza A and B in Macau, 2010–2018
2021, 36(5): 1144 doi: 10.1007/s12250-021-00388-6
Influenza is one of the major respiratory diseases in humans. Macau is a tourist city with high density of population and special population mobility. The study on the epidemiological characteristics of influenza in Macau should bring great value for preventing influenza in tourist cities like Macau in the world. In this study, we collected a total of 104,874 samples with influenza-like illness (ILI) in Macau from 2010 to 2018. Chi-square test and binary multivariable logistic regression were used to investigate the epidemiological characteristics of influenza A and B in Macau. Among these ILI samples, the overall positive rate is 17.17% for influenza A and 6.97% for influenza B. The epidemics of influenza in three years (i.e., 2012, 2017 and 2018) differ from the remaining years (i.e., normal years). In a normal year, influenza A occurs year-round whereas influenza B is seasonal. Our research shows significant differences in influenza infections between different age groups in normal years. Interestingly, our analysis shows no significant difference between locals and tourists in influenza A and B infection in a normal year, whereas the odds of influenza A in tourists were significantly higher than those in locals in July 2017 and the odds of influenza B in tourists were significantly higher than those in locals in January–February 2012 and January–February 2018. This is possibly attributed by the policy of free vaccination to everyone in Macau. These findings should be valuable for preventing influenza in not only Macau but also the world.
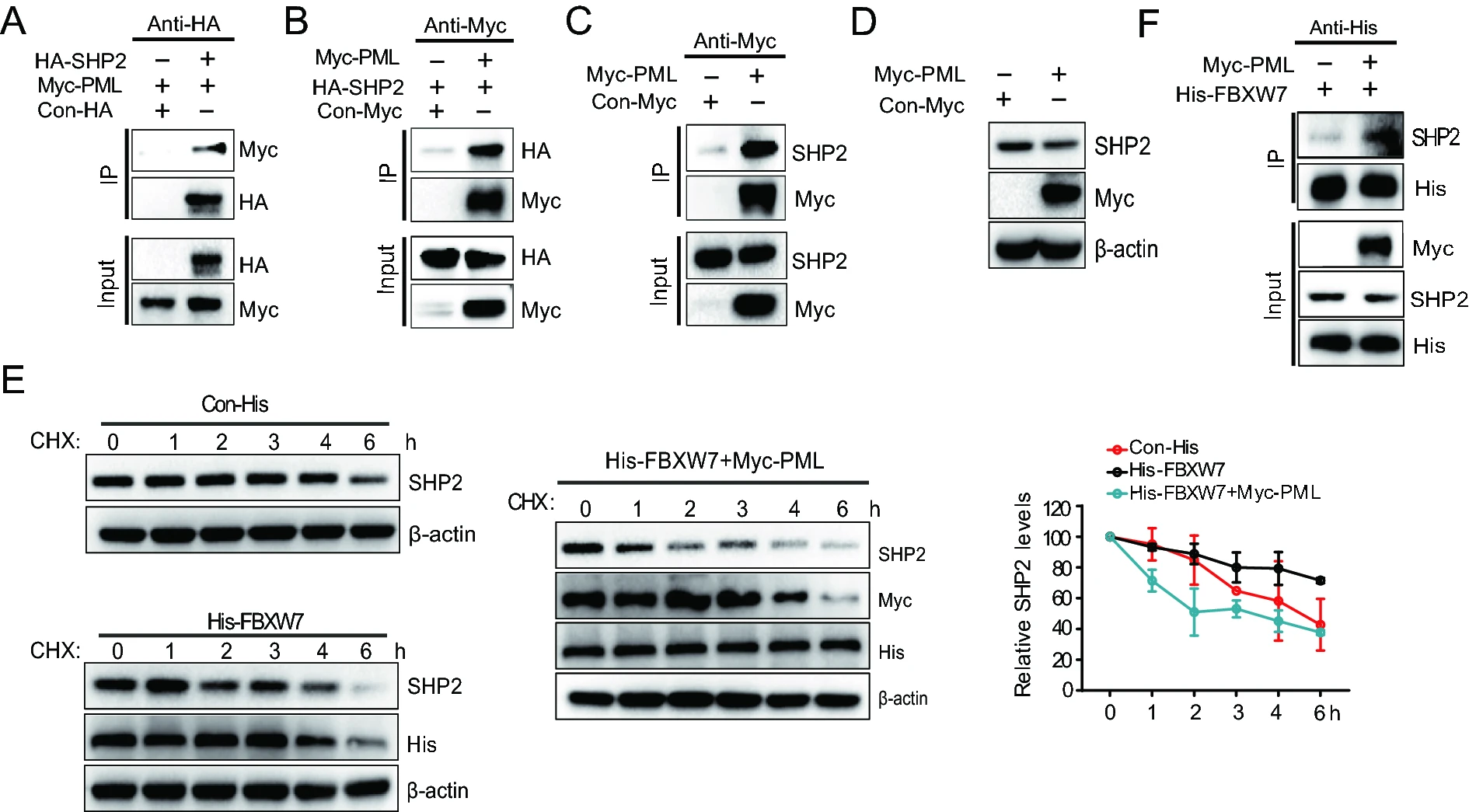
PML Suppresses Influenza Virus Replication by Promoting FBXW7 Expression
2021, 36(5): 1154 doi: 10.1007/s12250-021-00399-3
Influenza A viruses (IAV) are responsible for seasonal flu epidemics, which can lead to high morbidity and mortality each year. Like other viruses, influenza virus can hijack host cellular machinery for its replication. Host cells have evolved diverse cellular defense to resist the invasion of viruses. As the main components of promyelocytic leukemia protein nuclear bodies (PML-NBs), PML can inhibit the replication of many medically important viruses including IAV. However, the mechanism of PML against IAV is unclear. In the present study, we found PML was induced in response to IAV infection and ectopic expression of PML could inhibit IAV replication, whereas knockdown of endogenous PML expression could enhance IAV replication. Further studies showed that PML increased the expression of FBXW7 by inhibiting its K48-linked ubiquitination and enhanced the interaction between FBXW7 and SHP2, which negatively regulated IAV replication during infection. Moreover, PML stabilized RIG-I to promote the production of type Ⅰ IFN. Collectively, these data indicated that PML inhibited IAV replication by enhancing FBXW7 expression in the antiviral immunity against influenza virus and extended the mechanism of PML in antiviral immunity.
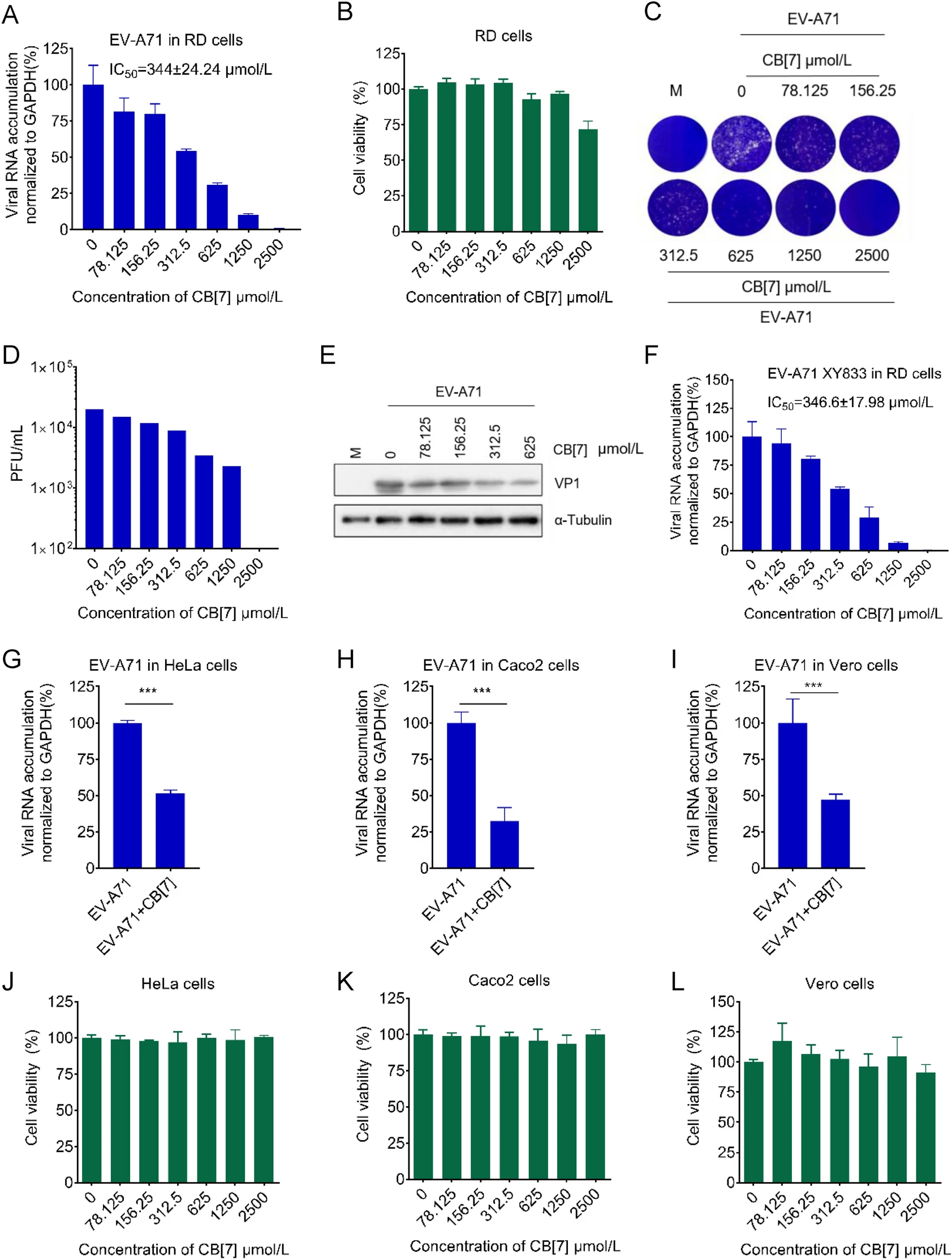
Cucurbit[7]uril as a Broad-Spectrum Antiviral Agent against Diverse RNA Viruses
2021, 36(5): 1165 doi: 10.1007/s12250-021-00404-9
The emergence and re-emergence of RNA virus outbreaks highlight the urgent need for the development of broad-spectrum antivirals. Polyamines are positively-charged small molecules required for the infectivity of a wide range of RNA viruses, therefore may become good antiviral targets. Cucurbit[7]uril (CB[7]), a synthetic macrocyclic molecule, which can bind with amine-based organic compounds with high affinity, has been shown to regulate bioactive molecules through competitive binding. In this study, we tested the antiviral activity of CB[7] against diverse RNA viruses, including a panel of enteroviruses (i.e. human enterovirus A71, coxsackievirus A16, coxsackievirus B3, and echovirus 11), some flaviviruses (i.e. dengue virus and Zika virus), and an alphavirus representative Semliki forest virus. CB[7] can inhibit virus replications in a variety of cell lines, and its mechanism of action is through the competitive binding with polyamines. Our findings not only for the first time provide evidence that CB[7] can be a promising broad-spectrum antiviral agent, but more importantly, offer a novel therapeutic strategy to fight against RNA viruses by supramolecular sequestration of polyamines.
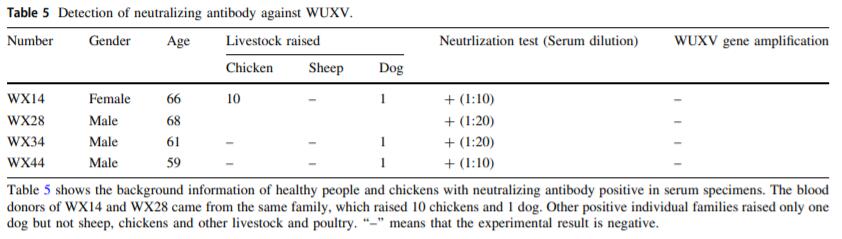
Re-isolation of Wuxiang Virus from Wild Sandflies Collected from Yangquan County, China
2021, 36(5): 1177 doi: 10.1007/s12250-021-00398-4
We previously isolated a new species of the genus Phlebovirus from wild sandflies collected from Wuxiang County in central China, which named the Wuxiang virus (WUXV). In this study, we re-isolated the WUXV from wild sandflies collected from two villages in Yangquan County, China in 2019. Four virus isolates that caused cytopathic effects in BHK-21 cells were successfully isolated from sandfly specimens collected from chicken pens and sheep pens. Phylogenetic analyses of the L, M and S gene segments of the viruses revealed that the four virus strains represented the previously isolated WUXV. The minimum infection rate (MIR) of the virus isolated from the sheep pen was 3.21, and the MIR of the virus isolated from the chicken pen was 3.45. The positive rates of Wuxiang virus neutralizing antibodies in serum samples of local healthy people and domestic chickens were 8.7% (4/46) and 100% (4/4), respectively, suggesting that Wuxiang virus can infect human and animal. In view of the fact that Wuxiang virus is infectious to humans and animals and has a relatively wide geographical distribution in China, it is of great public health significance to strengthen the investigation and study on the infection status of Wuxiang virus in humans and animals.
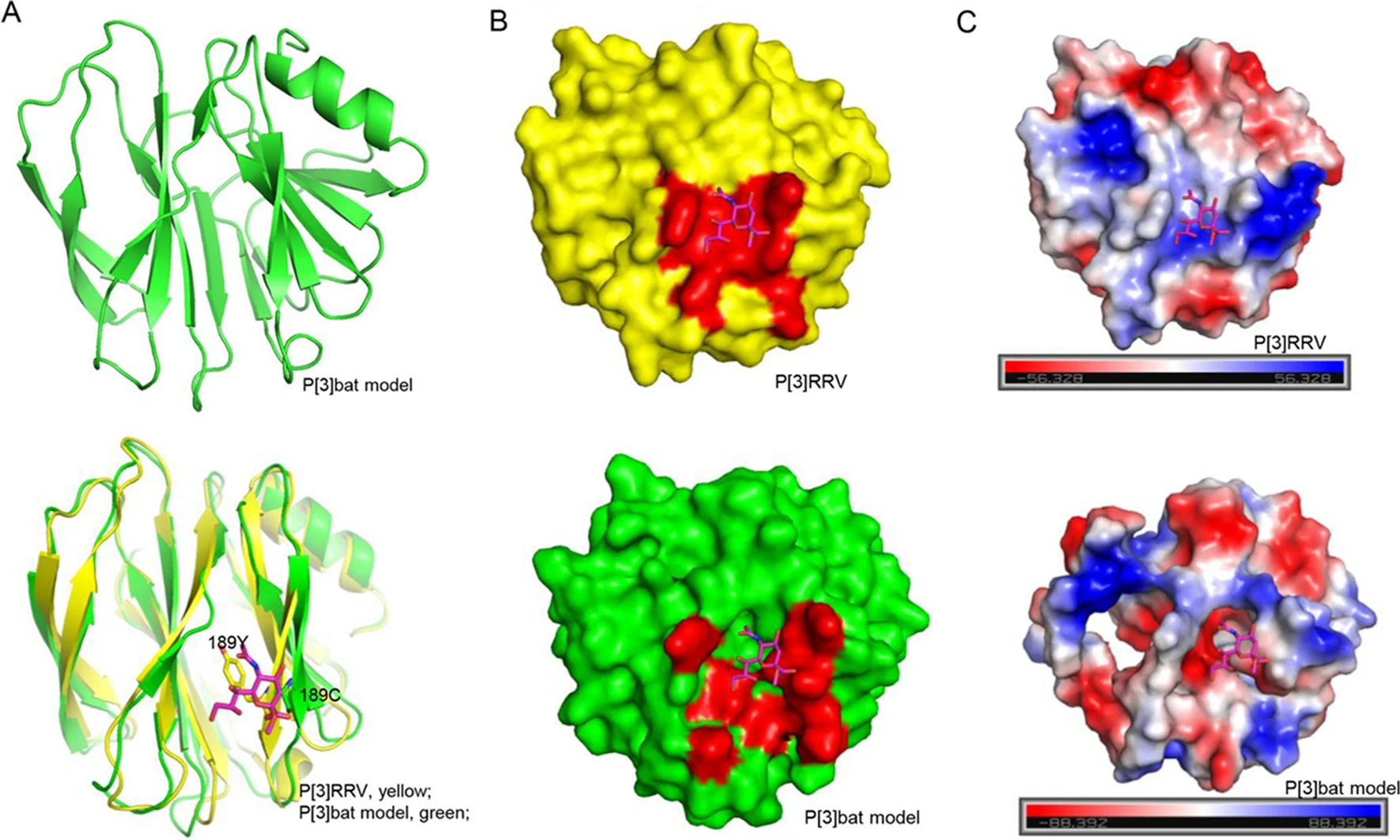
The Functional Characterization of Bat and Human P[3] Rotavirus VP8*s
2021, 36(5): 1187 doi: 10.1007/s12250-021-00400-z
P[3] rotavirus (RV) has been identified in many species, including human, simian, dog, and bat. Several glycans, including sialic acid, histo-blood group antigens (HBGAs) are reported as RV attachment factors. The glycan binding specificity of different P[3] RV VP8*s were investigated in this study. Human HCR3A and dog P[3] RV VP8*s recognized glycans with terminal sialic acid and hemagglutinated the red blood cells, while bat P[3] VP8* showed neither binding to glycans nor hemagglutination. However, the bat P[3] VP8* mutant of C189Y obtained the ability to hemagglutinate the red blood cells, while human P[3] HCR3A/M2-102 mutants of Y189C lost the ability. Sequence alignment and structural analysis indicated that residue 189 played an important role in the ligand recognition and may contribute to the cross-species transmission. Structural superimposition exhibited that bat P[3] VP8* model was quite different from the simian P[3] Rhesus rotavirus (RRV) P[3] VP8*, indicating that bat P[3] RV was relatively distinct and partially contributed to the no binding to tested glycans. These results promote our understanding of P[3] VP8*/glycans interactions and the potential transmission of bat/human P[3] RVs, offering more insight into the RV infection and prevalence.
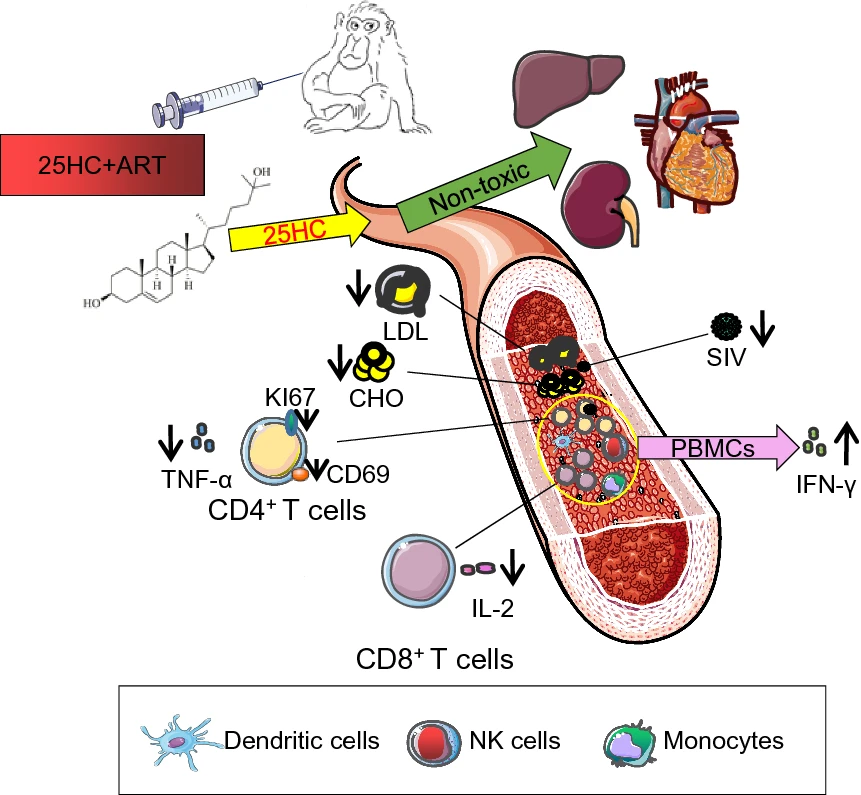
Modulation of Antiviral Immunity and Therapeutic Efficacy by 25-Hydroxycholesterol in Chronically SIV-Infected, ART-Treated Rhesus Macaques
2021, 36(5): 1197 doi: 10.1007/s12250-021-00407-6
Cholesterol-25-hydroxylase (CH25H) and its enzymatic product 25-hydroxycholesterol (25HC) exert broadly antiviral activity including inhibiting HIV-1 infection. However, their antiviral immunity and therapeutic efficacy in a nonhuman primate model are unknown. Here, we report that the regimen of 25HC combined with antiretroviral therapy (ART), provides profound immunological modulation towards inhibiting viral replication in chronically SIVmac239-infected rhesus macaques (RMs). Compared to the ART alone, this regimen more effectively controlled SIV replication, enhanced SIV-specific cellular immune responses, restored the ratio of CD4/CD8 cells, reversed the hyperactivation state of CD4+ T cells, and inhibited the secretion of proinflammatory cytokines by CD4+ and CD8+ T lymphocytes in chronically SIV-infected RMs. Furthermore, the in vivo safety and the preliminary pharmacokinetics of the 25HC compound were assessed in this RM model. Taken together, these assessments help explain the profound relationship between cholesterol metabolism, immune modulation, and antiviral activities by 25HC. These results provide insight for developing novel therapeutic drug candidates against HIV-1 infection and other related diseases.
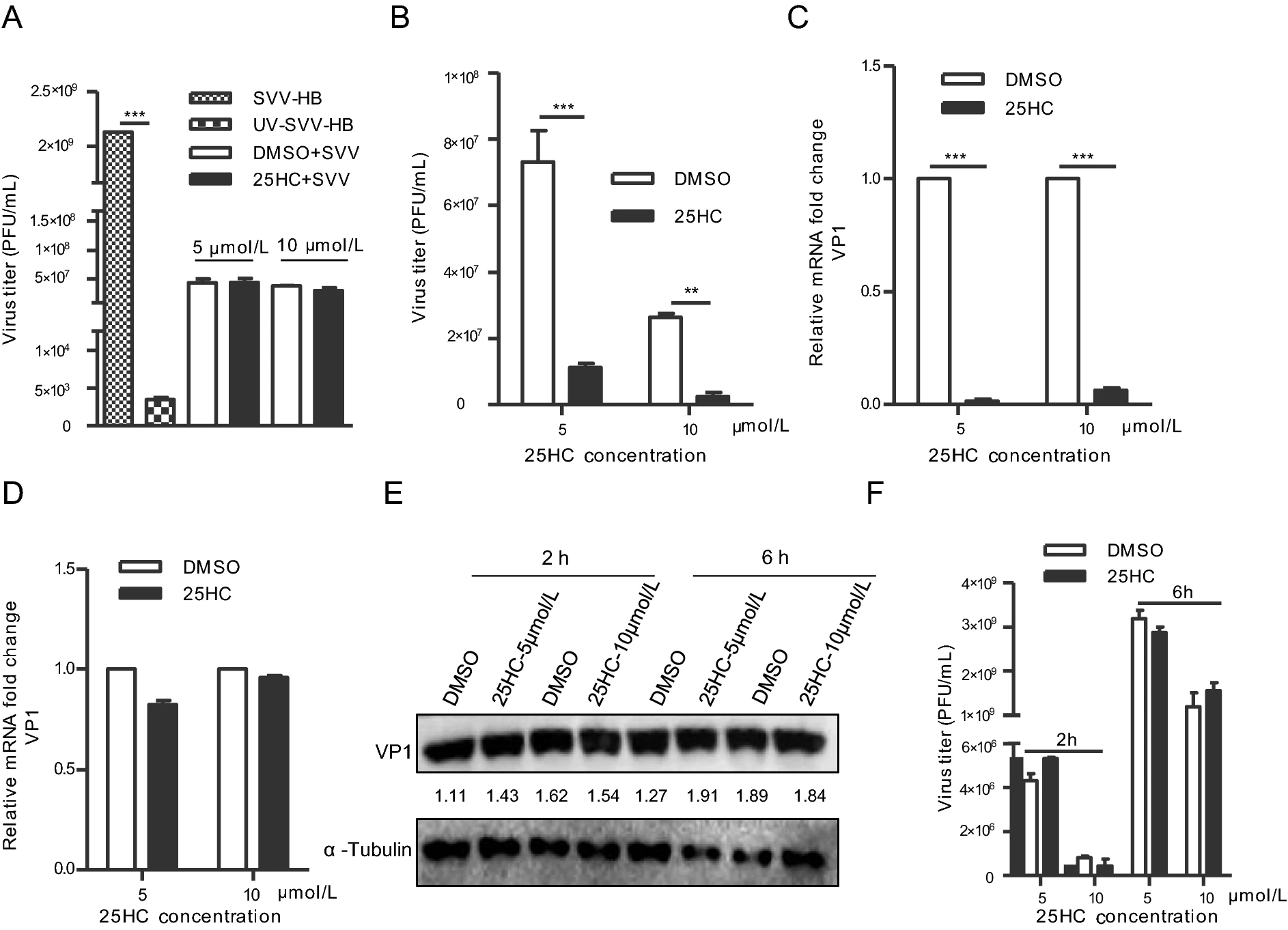
Cholesterol-25-Hydroxylase Suppresses Seneca Valley Virus Infection via Producing 25-Hydroxycholesterol to Block Adsorption Procedure
2021, 36(5): 1210 doi: 10.1007/s12250-021-00377-9
Cholesterol-25-hydroxylase (CH25H) is a membrane protein associated with endoplasmic reticulum, and it is an interferon-stimulated factor regulated by interferon. CH25H catalyzes cholesterol to produce 25-hydroxycholesterol (25HC) by adding a second hydroxyl to the 25th carbon atom of cholesterol. Recent studies have shown that both CH25H and 25HC could inhibit the replication of many viruses. In this study, we found that ectopic expression of CH25H in HEK-293T and BHK-21 cell lines could inhibit the replication of Seneca Valley virus (SVV) and that there was no species difference. On the other hand, the knockdown of CH25H could enhance the replication of SVV in HEK-293T and BHK-21 cells, indicating the importance of CH25H. To some extent, the CH25H mutant without hydroxylase activity also lost its ability to inhibit SVV amplification. Further studies demonstrated that 25HC was involved in the entire life cycle of SVV, especially in repressing its adsorption process. This study reveals that CH25H exerts the advantage of innate immunity mainly by producing 25HC to block virion adsorption.

Antigenic Drift of the Hemagglutinin from an Influenza A (H1N1) pdm09 Clinical Isolate Increases its Pathogenicity In Vitro
2021, 36(5): 1220 doi: 10.1007/s12250-021-00401-y
The influenza A (H1N1) pdm09 virus emerged in 2009 and has been continuously circulating in humans for over ten years. Here, we analyzed a clinical influenza A (H1N1) pdm09-infected patient case hospitalized for two months in Guangdong (from December 14, 2019 to February 15, 2020). This isolate, named A/Guangdong/LCF/2019 (LCF/19), was genetically sequenced, rescued by reverse genetics, and phylogenetically analyzed in the context of other relevant pdm09 isolates. Compared with earlier isolates, this pdm09 virus's genetic sequence contains four substitutions, S186P, T188I, D190A, and Q192E, of the hemagglutinin (HA) segment at position 186–192 (H3 numbering) in the epitope Sb, and two of which are located at the 190-helix. Phylogenetic analysis indicated that the epitope Sb started undergoing a rapid antigenic change in 2018. To characterize the pathogenicity of this novel substitution motif, a panel of reassortant viruses containing the LCF/2019 HA segment or the chimeric HA segment with the four substitutions were rescued. Kinetic growth data revealed that the reassortant viruses, including the LCF/2019 with the PTIAAQE substitution, propagated faster than those rescued ones having the STTADQQ motif in the epitope Sb in Madin-Darby Canine Kidney (MDCK) cells. The HI test showed that the binding activity of escape mutant to 2018 pdm09 sera was weaker than GLW/2018, suggesting that old vaccines might not effectively protect people from infection. Due to the difference in the selection of vaccine strains, people vaccinated in the southern hemisphere could still suffer a severe infection if infected with this antigenic drift pdm09 virus.
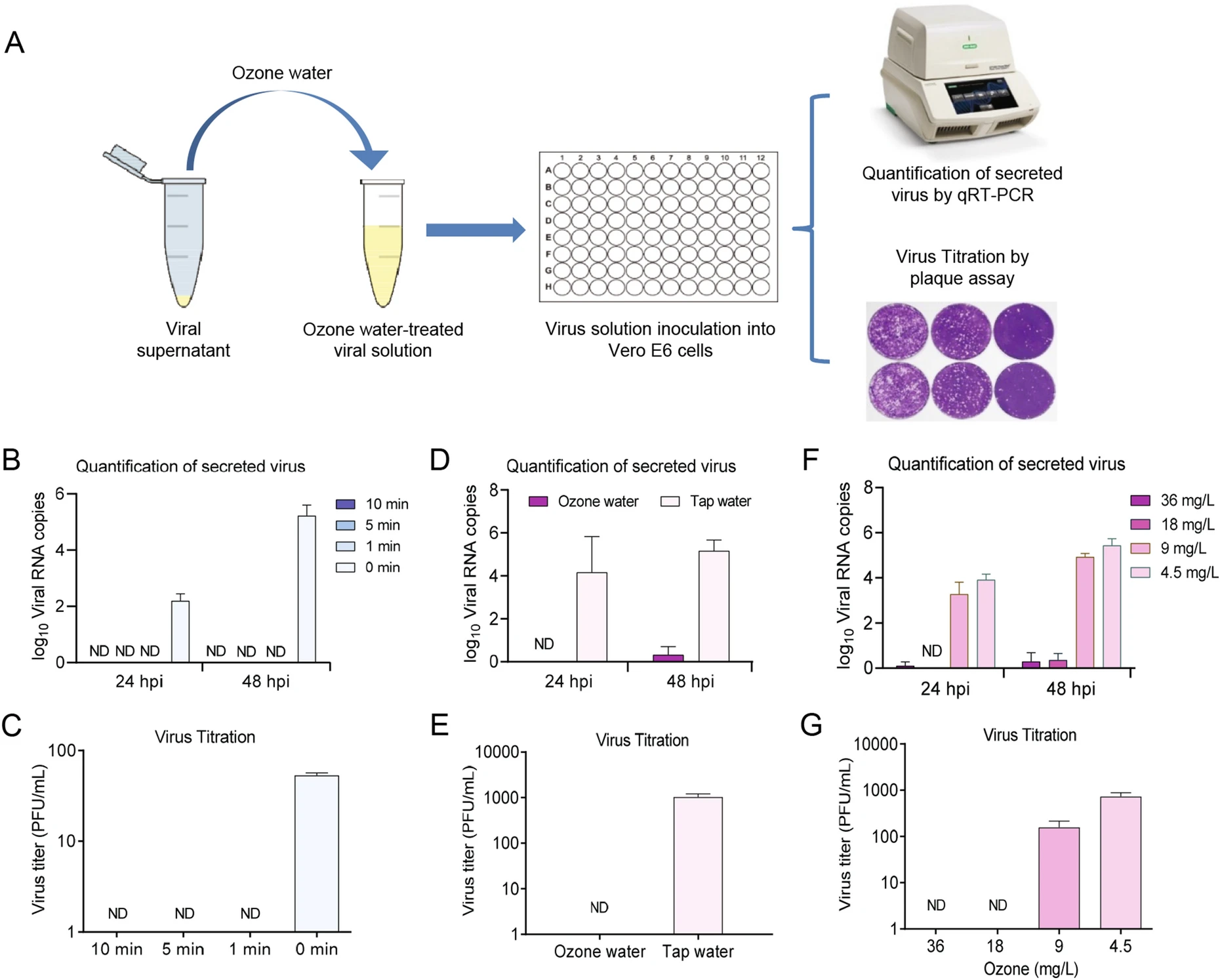
Ozone Water Is an Effective Disinfectant for SARS-CoV-2
2021, 36(5): 1066 doi: 10.1007/s12250-021-00379-7
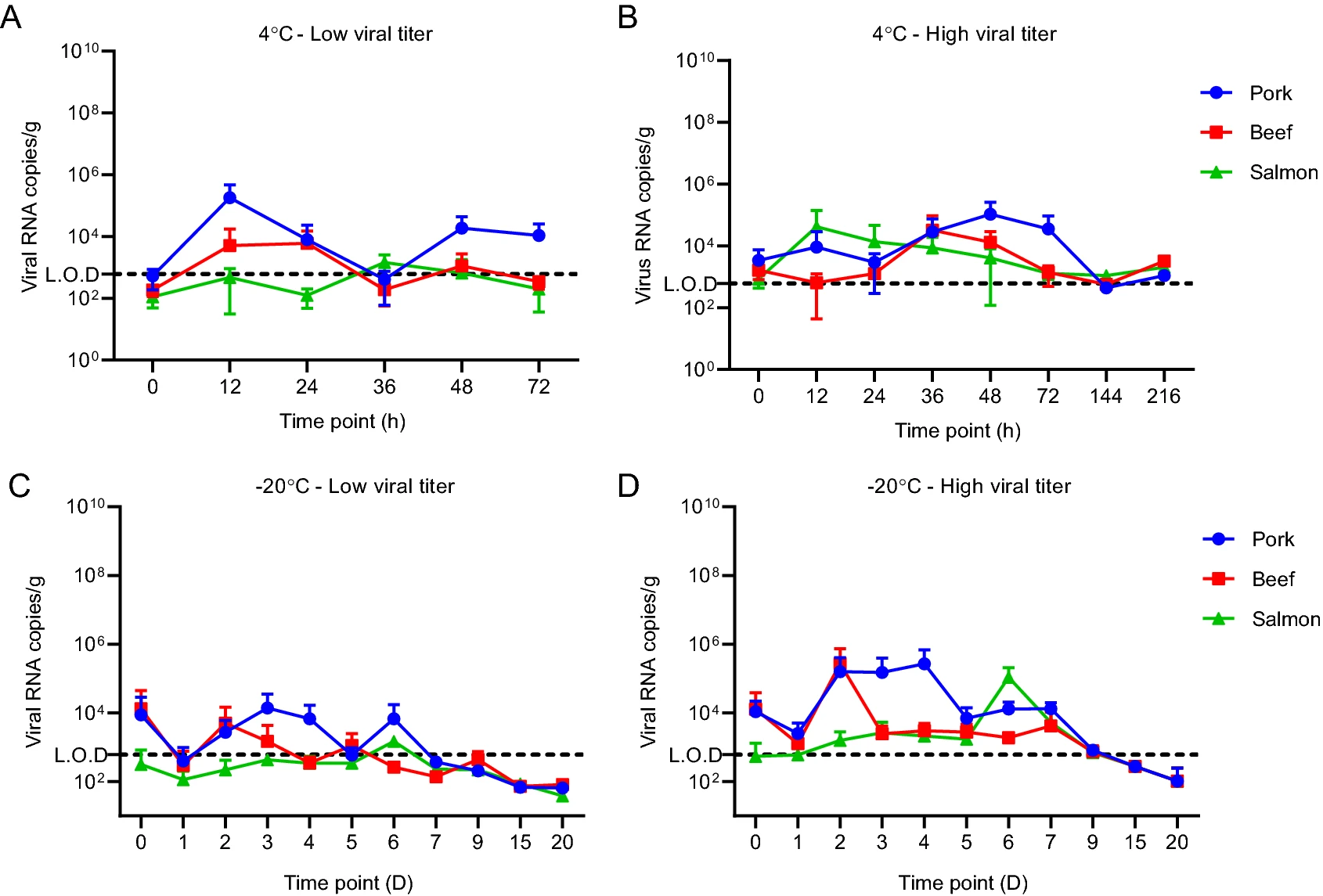
Stability of SARS-CoV-2 on the Surfaces of Three Meats in the Setting That Simulates the Cold Chain Transportation
2021, 36(5): 1069 doi: 10.1007/s12250-021-00367-x

Genetic Mutation of SARS-CoV-2 during Consecutive Passages in Permissive Cells
2021, 36(5): 1073 doi: 10.1007/s12250-021-00384-w
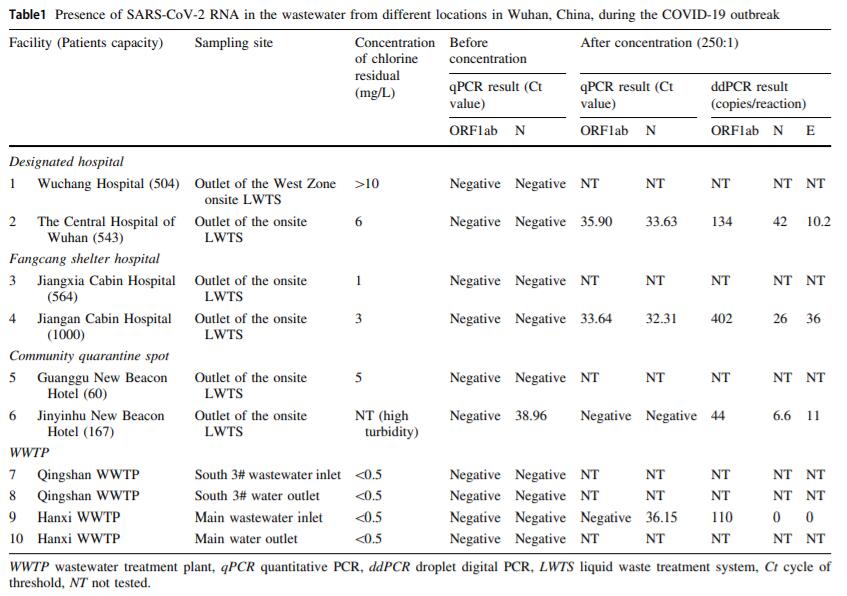
Detection of SARS-CoV-2 RNA in Medical Wastewater in Wuhan During the COVID-19 Outbreak
2021, 36(5): 1077 doi: 10.1007/s12250-021-00373-z
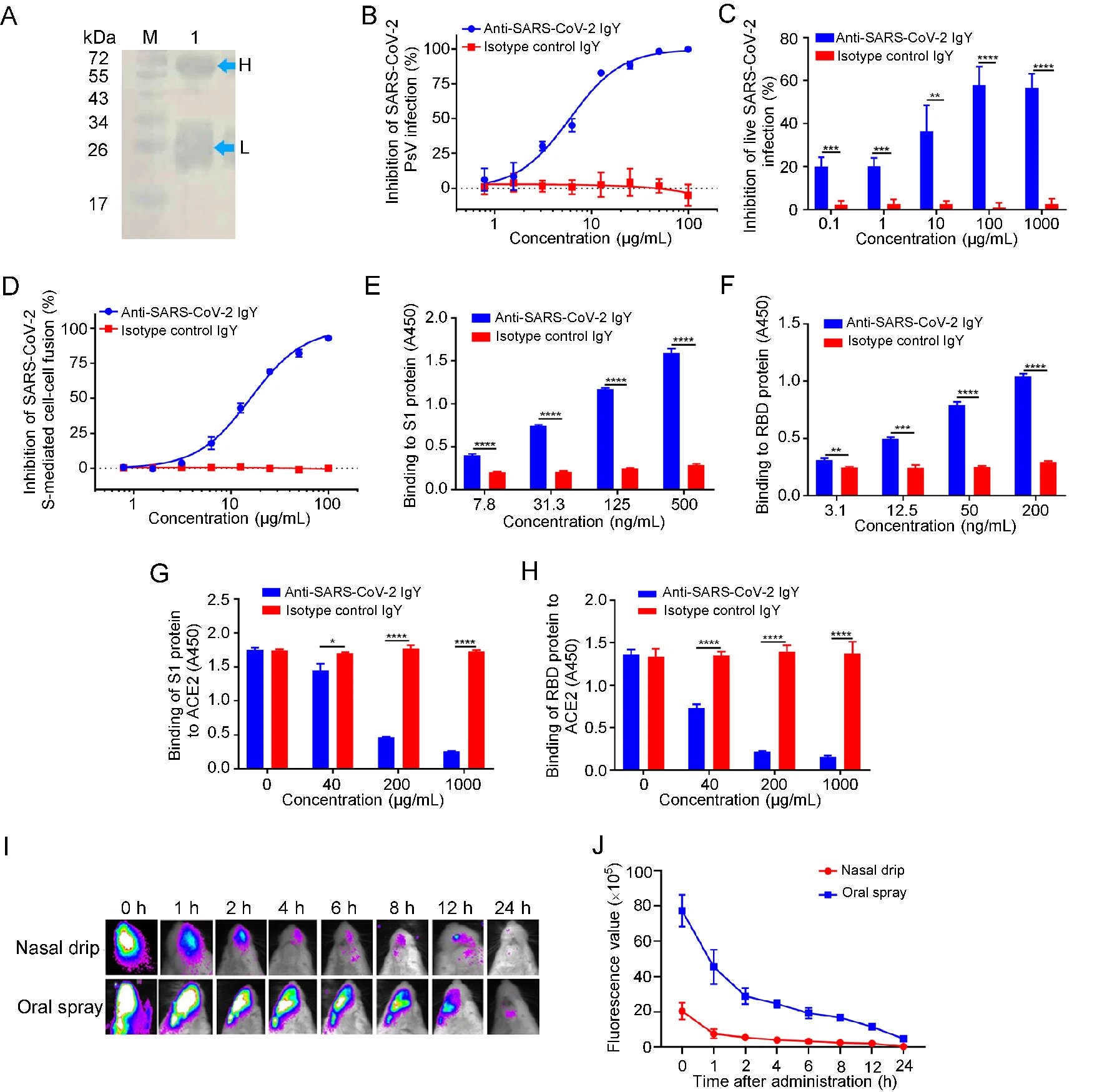
Anti-SARS-CoV-2 IgY Isolated from Egg Yolks of Hens Immunized with Inactivated SARS-CoV-2 for Immunoprophylaxis of COVID-19
2021, 36(5): 1080 doi: 10.1007/s12250-021-00371-1
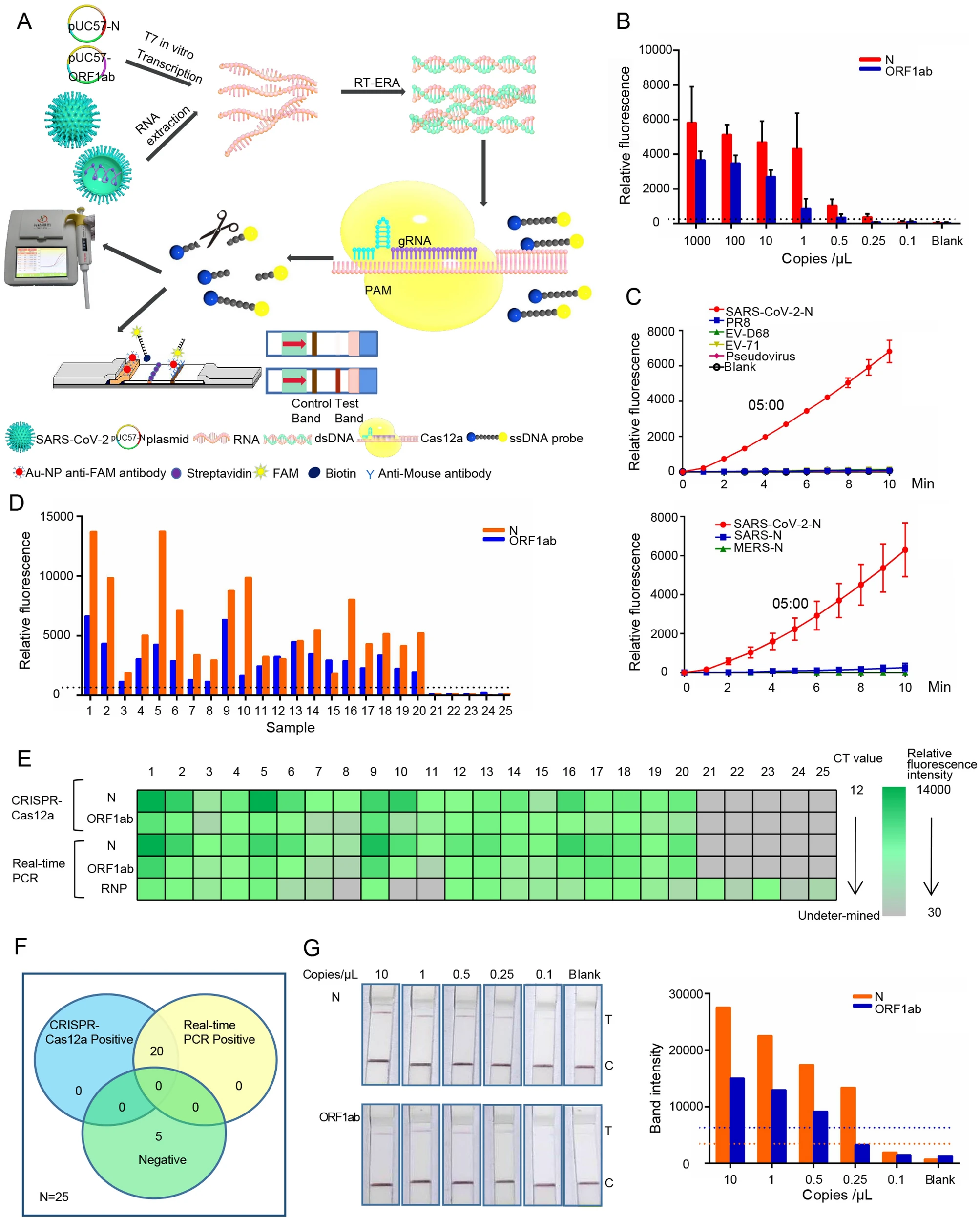
CRISPR/Cas12a Technology Combined with RT-ERA for Rapid and Portable SARS-CoV-2 Detection
2021, 36(5): 1083 doi: 10.1007/s12250-021-00406-7
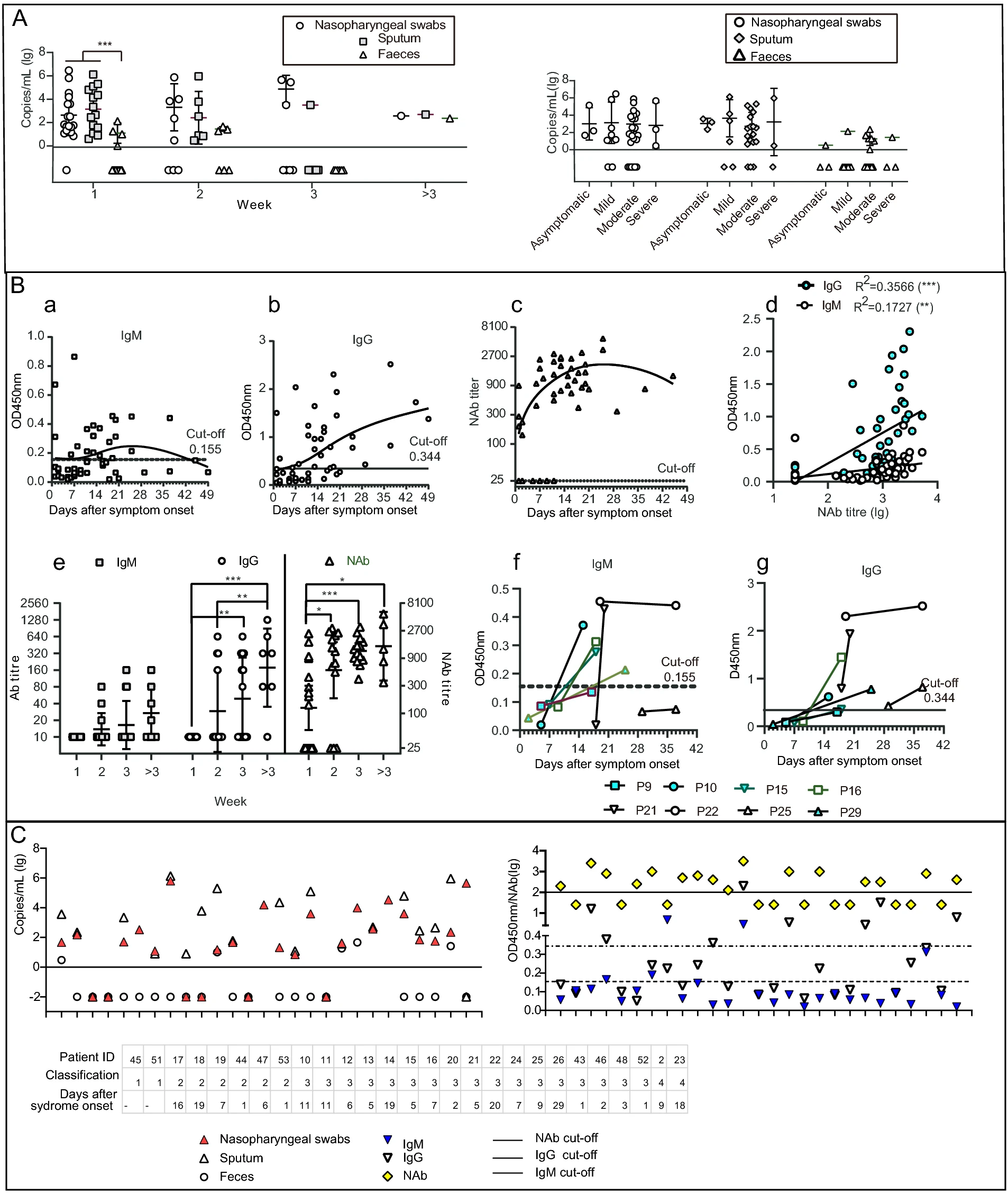
Profiles of SARS-CoV-2 RNA and Antibodies in Inpatients with COVID-19 not Related with Clinical Manifestation: A Single Centre Study
2021, 36(5): 1088 doi: 10.1007/s12250-021-00411-w
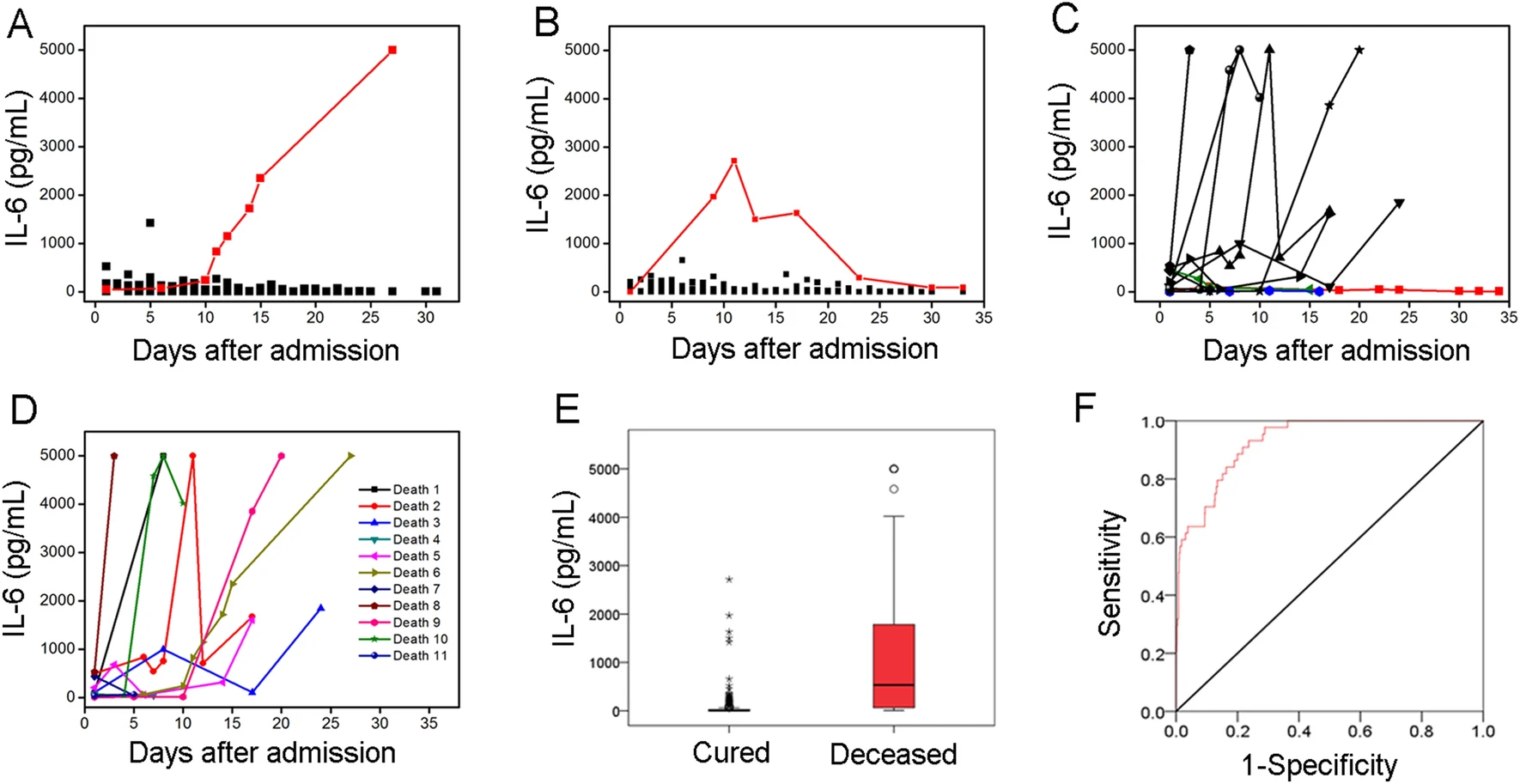
Consecutive Monitoring of Interleukin-6 Is Needed for COVID-19 Patients
2021, 36(5): 1093 doi: 10.1007/s12250-021-00425-4
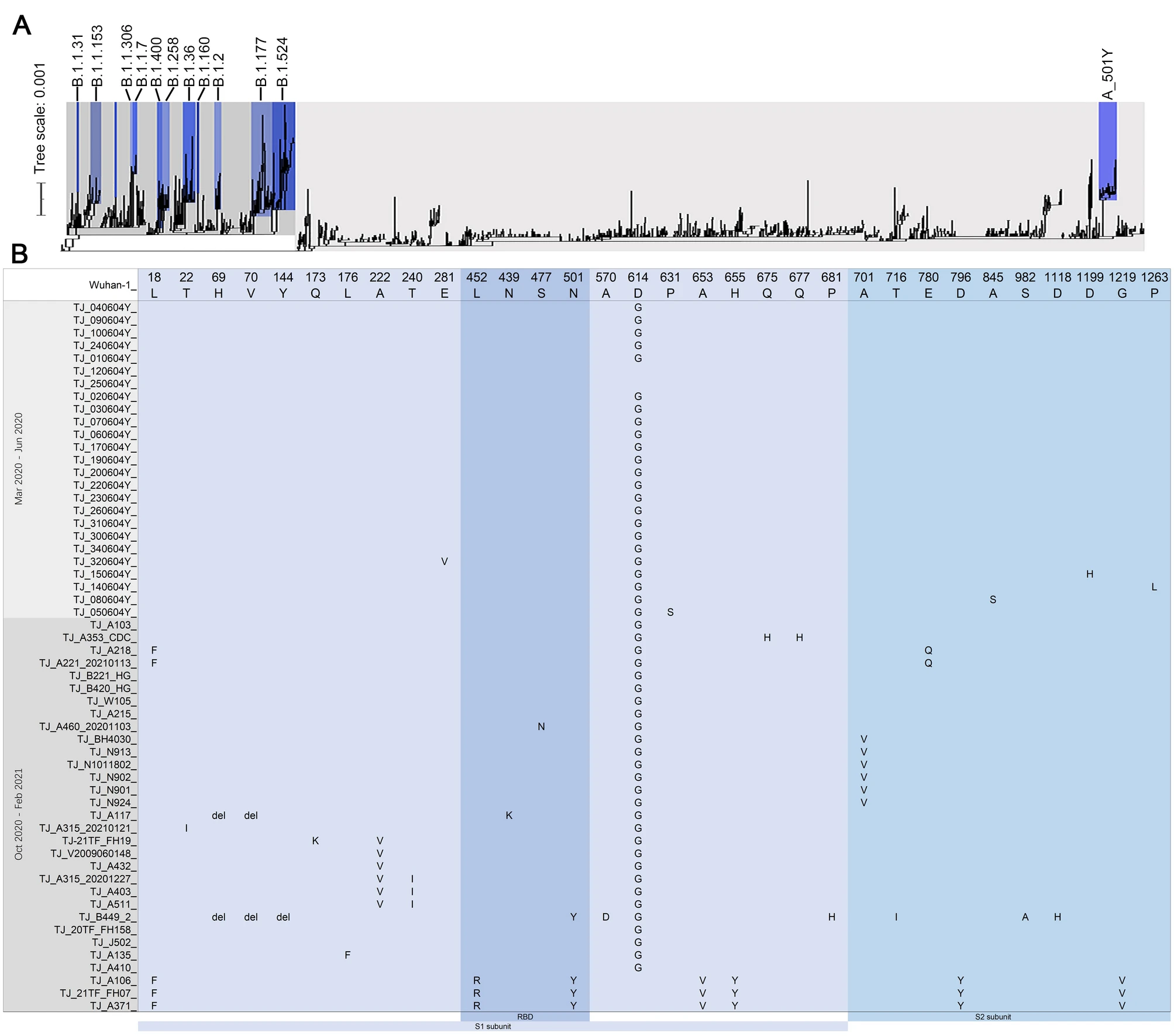
SARS-CoV-2 Genomic Sequencing Revealed N501Y and L452R Mutants of S/A Lineage in Tianjin Municipality, China
2021, 36(5): 1228 doi: 10.1007/s12250-021-00432-5
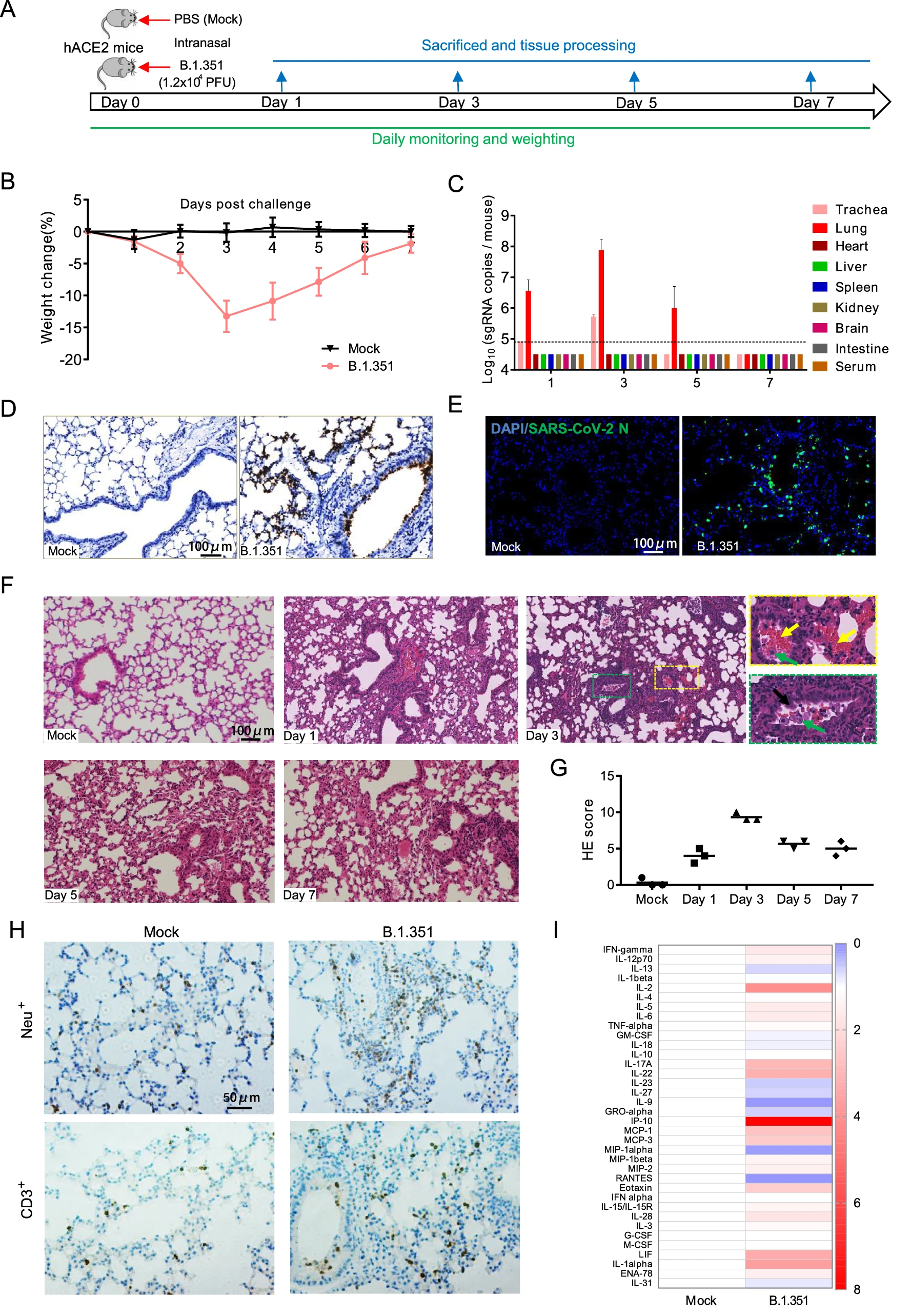
The Infection and Pathogenicity of SARS-CoV-2 Variant B.1.351 in hACE2 Mice
2021, 36(5): 1232 doi: 10.1007/s12250-021-00452-1
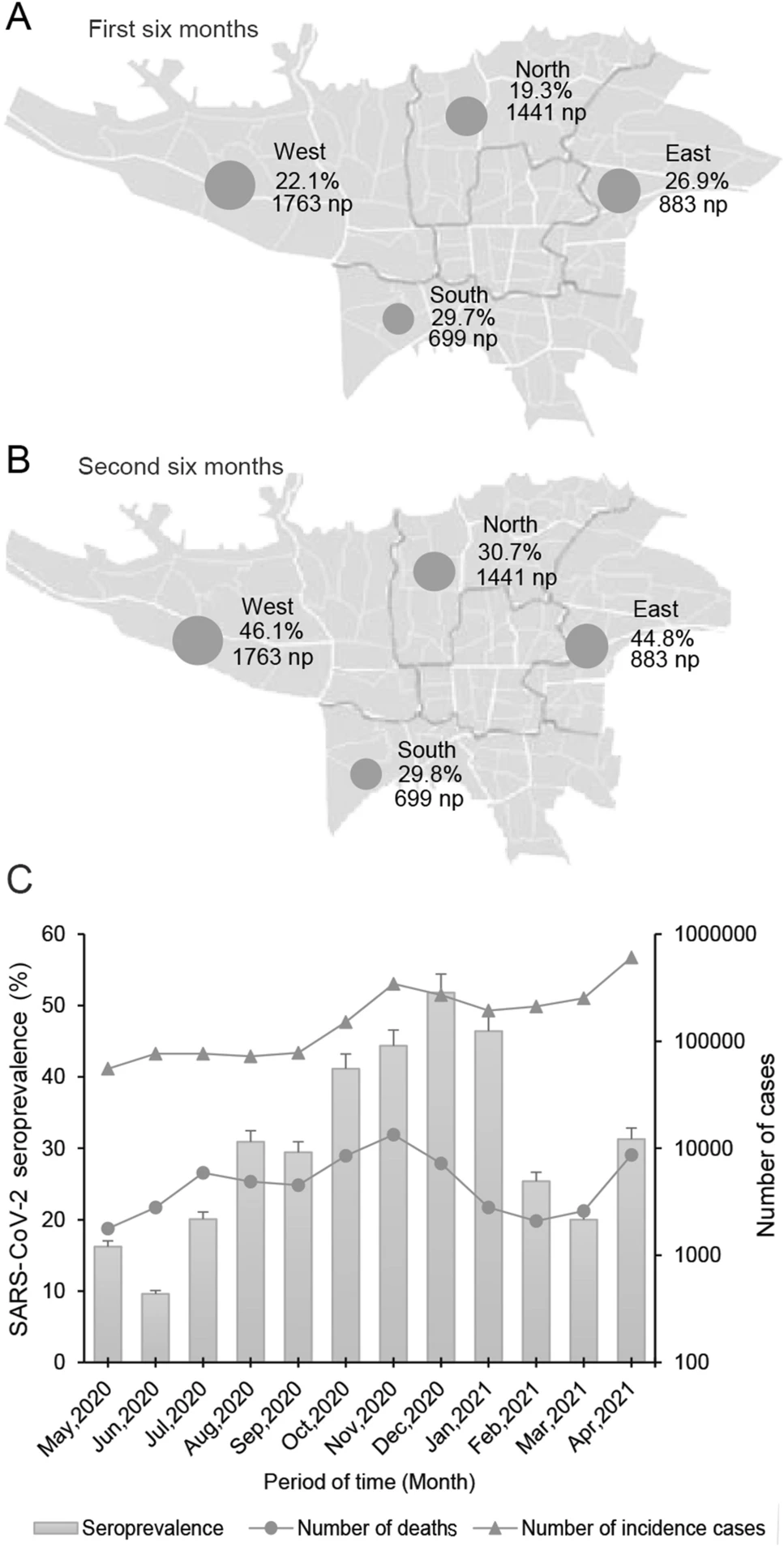
SARS-CoV-2 Seroprevalence in People Referred to Private Medical Laboratories in Different Districts of Tehran, Iran from May 2020 to April 2021
2021, 36(5): 1236 doi: 10.1007/s12250-021-00446-z
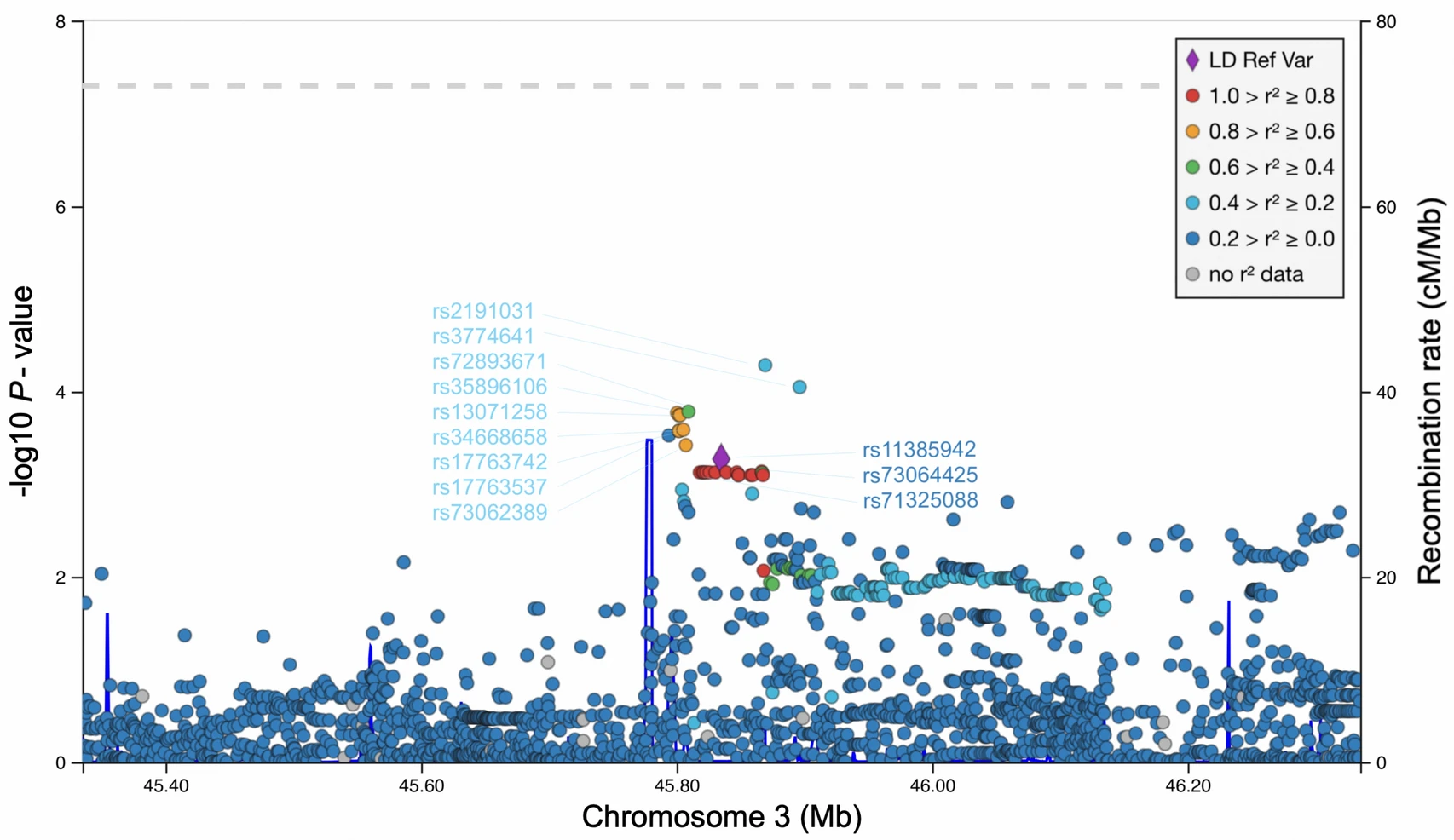
Replication of LZTFL1 Gene Region as a Susceptibility Locus for COVID-19 in Latvian Population
2021, 36(5): 1241 doi: 10.1007/s12250-021-00448-x

Recombinant GII.4[P31] Was Predominant Norovirus Circulating in Beijing Area, China, 2018–2020
2021, 36(5): 1245 doi: 10.1007/s12250-021-00381-z

Beagle Dogs Have Low Susceptibility to Florida Clade 2 H3N8 Equine Avian Influenza
2021, 36(5): 1248 doi: 10.1007/s12250-021-00366-y
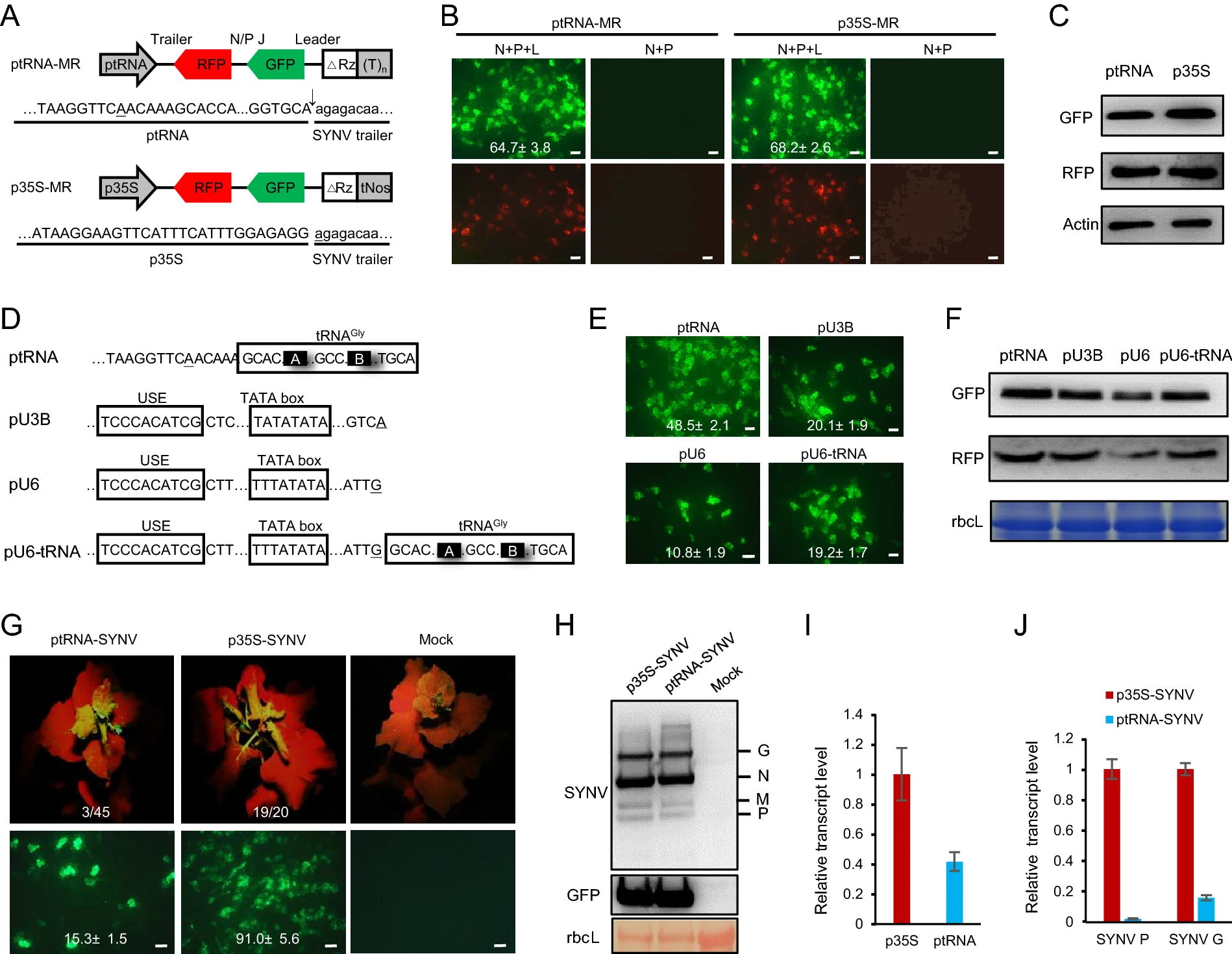
Development of RNA Polymerase III-Driven Reverse Genetics System for the Rescue of a Plant Rhabdovirus
2021, 36(5): 1252 doi: 10.1007/s12250-021-00390-y
Safety and Considerations of the COVID-19 Vaccine Massive Deployment
2021, 36(5): 1097 doi: 10.1007/s12250-021-00408-5
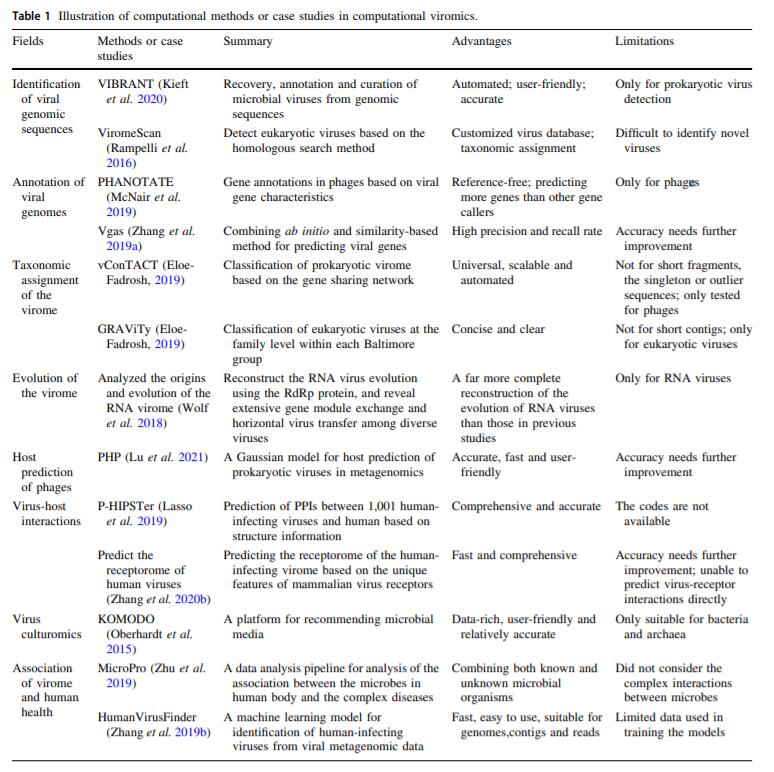
Computational Viromics: Applications of the Computational Biology in Viromics Studies
2021, 36(5): 1256 doi: 10.1007/s12250-021-00395-7

















