HTML
-
Phleboviruses belong to the Phenuiviridae family of the order Bunyavirales (Maes et al. 2019). In March 2020, the International Committee on Taxonomy of Viruses (ICTV) released the latest Phenuiviridae classification guidelines. The Phenuiviridae family includes 60 different virus types, such as the sandfly-borne Toscana virus (TOSV), Toros virus (TORV), the mosquito-borne Rift Valley fever virus, and sandfly fever Naples virus. According to the latest ICTV classification guidelines, the tick-borne severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus (HLV) are not included in the Phlebovirus genus (Kuhn et al. 2020; Walker et al. 2020). The number of newly identified Phleboviruses has increased drastically over the past few years, suggesting that Phleboviruses are abundant in nature.
In 2018, we isolated a sandfly-borne Wuxiang virus (WUXV) from a sandfly in China; we termed this isolate SXWX1813-2 virus (Wang et al. 2020). SXWX1813-2 virus inoculating in BHK-21 cells led to significant cytotoxicity. In newborn mice, the virus caused morbidity and death. Whole-genome sequencing of this new WUXV isolate revealed three gene segments (L gene, M gene, and S gene). Molecular genetic evolution analyses of the viral genome indicated that the WUXV differs phylogenetically from other Phleboviruses, forming an independent phylogenetic branch (Wang et al. 2020).
The SXWX1813-2 virus is the first Phlebovirus isolated from a sandfly in East Asia (Depaquit et al. 2010; Liang et al. 2018; Wang et al. 2020). However, it remains unclear whether there are sandfly-borne viruses in nature in other regions. In 2019, we isolated viruses from sandflies collected from Yangquan County, and identified four isolates that can cause cytopathic effects (CPE) in BHK-21 cells. Molecular analyses showed that all four isolates belonged to the WUXV. Herein, we report the re-isolation of a new Phlebovirus from a sandfly collected in a natural environment in China. The results also showed that local healthy people and chicken serum samples were positive for neutralizing antibody of Wuxiang virus.
-
Wild sandfly specimens were collected from chicken, sheep, and cattle pens used to breed blood-sucking insects. Specimens were collected using an insect collector (MM200; Guangzhou Changsheng Chemical Co., Ltd., Guangdong, China). The specimen collectors were placed at each sampling point from 6 PM to 7 AM. The collected insects were placed in a low-temperature refrigerator for 20 min and then kept on ice during morphological identification and isolation of blood-sucking insects. Sandfly specimens were divided based on the collection environment and stored in liquid nitrogen until further testing (Fu et al. 2017; Song et al. 2017; Wang et al. 2020).
Whole blood of local healthy people and chicken were also collected at place where the sandfly specimens were collected, and the sera were obtained after centrifugation at 2000 rpm for 20 min. Sera samples were transported to the laboratory with ice platoons, stored in a low temperature refrigerator for testing.
-
The golden hamster kidney cells BHK-21 were maintained in 90% Eagle's medium (made in-house) supplemented with 7% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), 1% penicillin and streptomycin (100 U/mL), 1% glutamine (30 g/L), and 1% NaHCO3. The Aedes albopictus egg cells C6/36 were cultured in 89% RMPI 1640 (Invitrogen) supplemented with 10% FBS (Invitrogen) and 1% penicillin and streptomycin (100 U/mL). BHK-21 and C6/36 cells were maintained in a 5% CO2 humified atmosphere. BHK-21 cells were cultured at 37 ℃, whereas C6/36 cells were kept at 28 ℃ (Fu et al. 2017; Song et al. 2017; Wang et al. 2020).
-
Sandfly specimens were pooled into 23 groups with 50-100 sandflies in each group. Specimens were placed in a glass grinder and homogenized in grinding solution (93% Eagle's medium, 5% penicillin and streptomycin [100 U/mL], 1% glutamine [30 g/L], and 1% NaHCO3) at 4 ℃. Samples were centrifuged (4 ℃, 12,000 rpm, 30 min), and 100 μL of the supernatant was inoculated into BHK-21 cell and C6/36 cells seeded at a density of 80% in 24-well plates (Corning Inc., New York, NY, USA). After inoculation, BHK-21 and C6/36 cells were cultured in a 5% CO2 atmosphere at 37 ℃ and 28 ℃, respectively. CPE was monitored under a microscope every 12 h. When CPE became evident, viral samples were collected and stored at -80 ℃ until further characterization. Specimens were blindly passaged three times; cells without CPE were discarded (Fu et al. 2017; Song et al. 2017).
-
Viral total RNA was extracted using a Viral RNA Mini Kit (QIAamp; Qiagen, Valencia, CA, USA) following the manufacturer's instructions. Total RNA was extracted from sandfly specimens after homogenization, as well as from the supernatant of infected BHK-21 and C6/36 cells. The extracted RNA was immediately placed in a 65 ℃ water bath for 10 min and then transferred on ice for 2 min. RNA samples (32 μL) were used for first-strand cDNA synthesis (Ready-To-Go You-Prime First-Strand Beads; GE Healthcare, Little Chalfont, UK). The reaction also included 1 μL of random primers (TaKaRa, Shiga, Japan) and was incubated at 37 ℃ for 1 h. The resulting cDNA was used immediately, or stored at -40 ℃ until further use (Feng et al. 2017; Ren et al. 2017; Wang et al. 2020).
-
Viral genes were amplified by polymerase chain reaction (PCR), which included cDNA template, GoTaq® Green Master Mix 2 × (Promega, Madison, WI, USA), and 10 μmol/L of each primer (total reaction volume of 25 μL). Amplification products were resolved by 1% agarose gel electrophoresis and subjected to sequencing (Feng et al. 2017; Ren et al. 2017; Wang et al. 2020). The sequences of the primers used in this study were previously described (Wang et al. 2020). We used nine primer pairs for WUXV M genes and six for WUXV S genes. Four M gene amplification primers and two S gene amplification primers did not yield amplification products. Therefore, amplification primers were redesigned for genes that could not be amplified. Specifically, we designed five M gene amplification primer pairs and two S gene amplification primer pairs. We designed eleven L gene primer pairs for PCR amplification. By using these primers, we obtained amplification products for all viral genes. The sequencing primers used in this study are listed in Supplementary Table S1.
-
BLAST analyses were conducted using the viral gene nucleotide sequences. SeqMan software (DNAStar, Madison, WI, USA) was used for sequence splicing and quality analyses, and BioEdit (version 7.0; http://www.mbio.ncsu.edu/BioEdit/bioedit.html) was used for multiple sequence alignment. System evolution analysis was conducted using MEGA 6.0 software (https://www.megasoftware.net/) and the neighbor-joining method (bootstrap value of 1000). MegAlign was used for homology analysis of nucleotide and amino acid sequences (DNAStar) (Feng et al. 2017, 2019; Song et al. 2017; Wang et al. 2020). Viral gene sequences were obtained from GenBank (NCBI, Bethesda, ND, USA) for comparison.
-
DNA extracted from pools of sandflies positive for WUXV was used for mitochondrial cytochrome C oxidase I (COI) amplification (Folmer et al. 1994) and sequencing. The sequences of the upstream COI primer (LCO1490; ggtcaacaaatcataaagatattgg) and downstream COI primer (HC02198; taaacttcagggtgaccaaaaaatca) were used for BLAST analyses of GenBank data to determine the sandfly species (Folmer et al. 1994; Wang et al. 2020).
-
The infection rate of a pool of positive specimens was calculated using the following formula: pool infection rate = number of positive specimens ÷ total number of specimens × 100%. Assuming that each positive pool contained only one infected sandfly, we calculated the MIR of 1000 sandflies as follows: number of positive specimen pools (number of infected sandflies) ÷ total number of sandflies × 1000 (Feng et al. 2012).
-
BHK-21 cells were introduced into a 6-well plate and grown into monolayer cells the next day, with a coverage rate of 80%. The viral solution was diluted with serum-free Eagle's solution to a concentration of 200 PFU/0.1 mL. Serum samples were placed in 56 ℃ water bath for 30 min to inactivate complement. Serum samples were diluted with serum-free Eagle's solution at 1:5. Set back titration of 100 PFU, 50 PFU, 10 PFU. The diluted virus solution was mixed with serum in equal proportion and incubated in 37 ℃ incubator for 1 h. 0.1 mL of the serum and virus mixture was added to each well, and the virus back titration was added to the wells at the same time for 2 wells per titer, and incubated at 37 ℃ for 1 h. Then 4 mL methylcellulose was added to cover cells per well, and the cells were cultured in incubator at 37 ℃. Cytopathy of cells was observed daily. When the plaques were obvious, crystal violet staining was performed, and plaques were observed and counted. The plaque reduction neutralization test (PRNT50) was used as the standard, when the corresponding plaque number was less than that of the standard, and the dilution ratio of serum samples was considered as the neutralization titer (Cao et al. 2016).
Sample Collection
Cells
Virus Isolation
Viral RNA Extraction and cDNA Synthesis
Gene Amplification and Sequencing
Nucleotide Sequence Analysis
Molecular Identification of Sandflies
Minimum Infection Rate (MIR)
Detection of Neutralizing Antibody
-
Sandfly specimens were collected in June 2019 from sheep, chicken, and cattle pens in two villages in Yangquan County, Shanxi Province. A total of 1535 sandflies were collected from the two villages (1047 and 488 from the first and second villages, respectively). In total, 935 of these sandflies were collected from a sheep pen, 290 from a chicken pen, and 310 from a cattle pen (Table 1).
Collection site Breeding places Total Sheep pen Chicken pen Cattle pen 1 554 183 310 1047 2 381 107 / 488 Total 935 290 310 1535 "/" represents the missing cattle pen.
"1" indicates the first sampling village (east longitude 113°37′12″, north latitude 37°46′48″). There were more than 100 chickens in the chicken pen, 170 sheep in the sheep pen, and 18 cattle in the cattle pen. The distance between the three pens was less than 1000 m. "2" indicates the second sampling village (east longitude 113°38′24″, north latitude 37°45′). There were 70 sheep in the sheep pen and 10 chickens in the chicken pen. Sheep and chicken pens were in the courtyard of the residents, which had two dogs in the house. The two pens were approximately 2 km apart, and the two villages were about 4 km apart.Table 1. Collection sites of sandfly specimens in Yangquan County, Shanxi Province in 2019.
-
The collected sandflies were pooled into 23 groups before homogenization, and the supernatant of each pool was inoculated into BHK-21 and C6/36 cells after homogenization. SXYQ1916 sandfly specimens were also inoculated into BHK-21 cells, and CPE was observed after 3 days of inoculation. Notably, cells exhibited significant shrinkage and shedding (Fig. 1). Sandfly specimens were inoculated into BHK-21 cells, and continued culture and passaging of cells, then four virus strains were isolated (Table 2). In contrast, the inoculation of sandfly supernatants into C6/36 cells did not lead to CPE. And we obtained nothing after PCR amplification using C6/36 cell supernatants and WUXV-specific primers.

Figure 1. CPE of the WUXV isolate SXYQ1916 in BHK-21 cells. A BHK-21 cells as negative control were cultured for 3 days. B BHK-21 cells were also cultured for 3 days after the inoculation of WUXV isolate SXYQ1916. Culture in the presence of the SXYQ1916 virus led to a decrease in the number of adherent BHK-21 cells, and profound cell rounding and detachment. Magnification, ×100.
Strain number Collection site Breeding places Number of Phlebotomus chinensiss CPE/ Gene amplification BHK-21 cell WUXV C6/36 cell WUXV L M S M S SXYQ1931 1 Sheep pen 60 + / MW192757 MW192753 – – – SXYQ1916 2 Sheep pen 49 + / MW192754 MW192750 – – – SXYQ1930 2 Sheep pen 74 + / MW192756 MW192752 – – – SXYQ1927-2 2 Chicken pen 54 + MW368897 MW192755 MW192751 – – – Numbers indicate the collection location. Numbers 1 and 2 indicate the first and second sampling villages, respectively; " + " means that the experimental result is positive; "–" means that the experimental result is negative; "/" means that the experiment was not performed. Table 2. Isolation of Wuxiang virus in Yangquan County, Shanxi Province in 2019.
-
By dividing all 1535 sandflies into 23 pools and inoculating them into BHK-21 cells, we obtained four virus isolates that caused CPE in BHK-21 cells. The infection rate of these viral pools was 17.39% (4/23), and the MIR was 2.61 (4/1535 × 1000). Among the 14 pools of sandfly specimens collected from the sheep pen, three contained the WUXV. The infection rate of WUXV pools isolated from the sheep pen was 21.43% (3/14), and the MIR was 3.21 (3/935 × 1000). The infection rate of viral pools isolated from the chicken pen was 20.00% (1/5), and the MIR of the WUXV virus was 3.45 (1/290 × 1000). In addition, 310 sandflies were collected from the cattle pen and divided into four pools. We could not isolate a virus from any of these viral pools, and we did not detect any of the WUXV genes. The infection rates of the different WUXV isolates are shown in Table 3.
Breeding places Number of sandflies Pool Infection rate of pools (%) MIR (/1000) Chicken pen 290 5 20.00 (1/5) 3.45 (1/290) Sheep pen 935 14 21.43 (3/14) 3.21 (3/935) Cattle pen 310 4 0.00 (0/4) 0.00 (0/310) Table 3. Infection rate of Sandfly-borne virus.
-
The sequences of the coding regions of the virus WUXV S and M genes were determined for all four virus isolates. Interestingly, the two genes were of the same length in all four virus isolates. The coding sequence of the S gene was 1611 base pairs (bp), whereas the coding sequence of the M gene was 4089 bp, similar to the length of the respective genes of the WUXV (SXWX1813-2). The L gene of the SXYQ1927-2 virus isolate was amplified, and the nucleotide sequence was determined and analyzed. The length of the nucleotide and the amino acid sequences of the encoding region of the L gene are 6273 nt and 2090 aa, respectively. BLAST analyses revealed that the sequences of the M gene exhibited similarities of 96.8%-99.5% among the four strains. Similarly, the amino acid sequences of the NS and N proteins encoded by the S gene were 98.2%-99.6% similar among the four strains (Supplementary Tables S2-S4). The results of the homology analysis of the newly isolated virus (SXYQ 1916) and other Phleboviruses are shown in Table 4. We found 96.7%, 97.1%, and 98.0% sequence similarity in genes M, NS, and N between the newly isolated strain SXYQ1916 and a WUXV strain (SXWX1813-2) previously isolated in China (Wang et al. 2020); the respective amino acid sequence similarities were 97.6%, 97.7%, and 99.6%. Moreover, comparisons between SXYQ1916 and a TORV strain (213/Turkey/2012) isolated from Turkey sandfly specimens revealed 72.0% nucleotide (75.1% amino acid) sequence similarity in the M segment, 75.4% (85.1%) similarity of the NS protein in the S segment, and 82.2% (96.4%) similarity of the N protein in the S segment (Table 4). These results suggested that the virus isolated from sandflies in Yangquan County is the same with the WUXV strain previously isolated from sandflies in China.
Virus strains M segment S segment GP NS N nt (%) aa (%) nt (%) aa (%) nt (%) aa (%) SXYQ1916 4089 1362 783 260 741 246 SXWX1813-2 4089(96.7) 1362(97.6) 783(97.1) 260(97.7) 741(98.0) 246(99.6) TORV(213/Turkey/2012) 4080(72.0) 1359(75.1) 783(75.4) 260(85.1) 741(82.2) 246(96.4) TORV(292/Turkey/2012) 4081(72.0) 1359(75.1) 783(75.2) 260(85.1) 741(81.9) 246(96.0) CFUV(Pa Ar 814/Greece/1981) 4080(71.3) 1359(75.7) 783(74.5) 260(84.3) 741(81.9) 246(96.4) SFSV(Ethiopia-2011/Ethiopia / 2011) 4026(62.6) 1341(57.1) / / 741(74.4) 246(84.2) SFSV(Sabin/Italy/1943) / / 783(53.3) 260(63.6) 741(75.3) 246(83.8) DASHV(131/Iran/2011) 4029(65.2) 1342(58.6) 786(58.1) 261(61.7) 741(76.5) 246(85.0) RVFV(ZH-548/Egypt/1977) 3594(36.0) 1197(39.9) 798(15.5) 265(24.1) 738(55.4) 245(52.8) "/" indicates that the sequence was not available in the GenBank database. Table 4. Homology analysis of the newly isolated virus (SXYQ 1916) and other Phleboviruses.
-
We performed phylogenetic analysis based on the genetic sequences of the four virus strains isolated from sandfly specimens in Yangquan County in 2019, and those of 60 sandfly viruses reclassified by ICTV in 2020. We found that the four virus strains isolated in this study belonged to the same evolutionary branch as the previously isolated WUXV strain (SXWX1813-2). On the basis of the sequences of the S (including the NS and N coding sequences), M and L gene, these viruses belonged to the same evolutionary population as the TORV isolated from Turkey sandflies in 2012-2013 (Fig. 2).
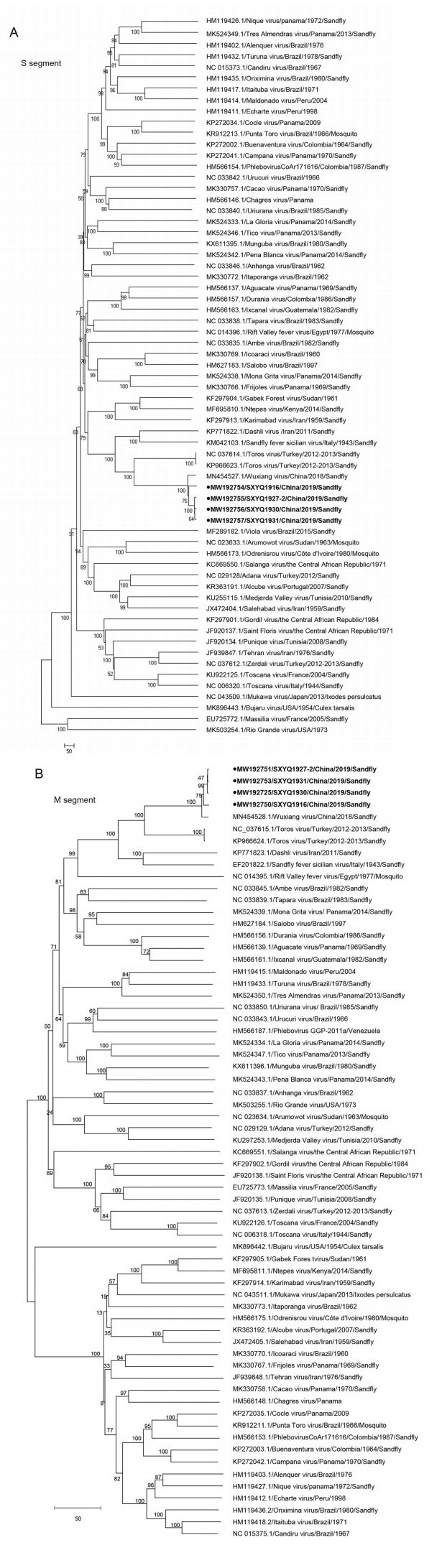
Figure 2. Phylogenetic analyses of the genetic sequences of the four virus strains isolated from sandflies collected in Yangquan in 2019. A Phylogenetic analysis of isolates SXYQ1916, SXYQ1927-2, SXYQ1930, SXYQ1931 (black dots) based on the M gene sequence. B Phylogenetic analysis of isolates SXYQ1916, SXYQ1927-2, SXYQ1930, SXYQ1931 based on the S gene sequence (black dots). C Phylogenetic analysis of isolates SXYQ1927-2 (black dots) based on the L gene sequence. The phylogenetic analysis was conducted using MEGA 6.0 software and the neighbor-joining method (bootstrap value of 1000). Note For phylogenetic comparison, we used the gene sequences of Phleboviruses from the ICTV 2020 guidelines; "/" indicates that the sequence was not available in the GenBank database.
-
To identify the species of the collected sandflies, we used PCR to amplify the COI gene. We found that all four pools of sandflies used to isolate the viruses belonged to the species Phlebotomus chinensis (Table 2).
-
In this study, 46 serum samples of healthy people were collected. The neutralizing antibody level of WUXV in serum samples was detected by plaque neutralization test (when the ratio of WUXV and neutralizing antibody in serum was 1:10, the sample was positive). The results showed that 8.70% (4/46) of the serum samples were positive in local heathy people. The neutralization titers of WX14, WX44 positive specimens were 1:10, WX28 and WX34 positive specimens were 1:20. The serum neutralizing antibodies of four chickens were all positive with titers ranging from 1:10 to 1:160. The results of neutralizing antibody test were shown in Table 5.
Number Gender Age Livestock raised Neutrlization test (Serum dilution) WUXV gene amplification Chicken Sheep Dog WX14 Female 66 10 – 1 + (1:10) – WX28 Male 68 + (1:20) – WX34 Male 61 – – 1 + (1:20) – WX44 Male 59 – – 1 + (1:10) – Table 5 shows the background information of healthy people and chickens with neutralizing antibody positive in serum specimens. The blood donors of WX14 and WX28 came from the same family, which raised 10 chickens and 1 dog. Other positive individual families raised only one dog but not sheep, chickens and other livestock and poultry. "–" means that the experimental result is negative. Table 5. Detection of neutralizing antibody against WUXV.
Sandfly Sample Collection
Virus Isolation
The MIR of WUXV
Homology in the Gene Sequences of Virus Isolates
Phylogenetic Analysis of Viral Genes
Identification of Sandfly Species
Detection of Neutralizing Antibody of WUXV
-
We have previously isolated the sandfly-borne virus WUXV from wild sandflies collected from Wuxiang County, Shanxi Province, China (Wang et al. 2020). In this study, we re-isolated WUXV from wild sandflies collected from Yangquan County, Shanxi Province, China. Molecular phylogenetic analyses revealed that these virus isolates represented different geographical isolates of WUXV viruses, suggesting that the WUXV can also be found in wild sandflies outside Wuxiang County.
Yangquan County (east longitude 112°5′-114°4′, north latitude 37°40′-38°31′) is located in Shanxi Province in central China, on the west side of the central Taihang Mountains with an average elevation of 700-1700 m. Yangquan has a warm, arid continental climate characterized by limited rainfall, and an ecological environment favoring the growth and reproduction of blood-sucking insects, such as sandflies and mosquitoes. In this study, we collected sandfly specimens from two regions of Yangquan County. One sampling point was located 9 km southeast of the center of Yangquan town, and the other was 13 km from the town; the distance between the two specimen collection points was approximately 4 km. The farmland around the two villages was mainly planted with corn and sorghum. Specimens were collected from sheep and chicken pens at each of the two villages. From the first village, we also collected samples from a cattle pen. All specimens were collected on June 11, 2019. Specimen collectors were placed at two collection points at the same time, and each of the sheep and chicken pens had one collector. Collectors were left overnight, and a total of 1535 sandflies were collected from both villages. Most of the sandflies were collected from the sheep pens (n = 935); only 290 samples were from the chicken pens. Three (SXYQ1916, SXYQ1930, SXYQ1927-2) of the four virus isolates in this study were derived from the sheep pen, and one isolate (SXYQ 1931) came from the chicken pen. These results demonstrate that sandflies were abundant in Yangquan County in June 2019.
Four different virus strains were isolated from wild sandflies collected from Yangquan County. The infection rate of these virus isolates differed significantly. The infection rate of the WUXV pool from the sheep pen was 21.43%, and the MIR was 3.21; the infection rate of the viral pool from the chicken pen was 20.00%, and the MIR was 3.45. In previous studies conducted in coastal countries in the Mediterranean region, the MIR of the TOSV isolated from Turkey sandflies was 2.2 (Calzolari et al. 2018), the MIR of Ntepes virus isolated from Kenyan sandflies was 0.26 (Tchouassi et al. 2019), and the MIR of Zerdali virus isolated from Turkey sandflies was 0.35 (Alkan et al. 2016). TORV was isolated from sandflies collected from Turkey in 2012, and its MIR was 0.26 (Alkan et al. 2016). Although phylogenetic analyses revealed that the WUXV isolated in Yangquan County, China, and the TORV isolated from Turkey were closely related, the MIR of the WUXV was significantly higher than that of the TORV (2.61 vs. 0.26). The pooled infection rate and MIR of WUXV from local sandflies were significantly higher than those of viruses isolated from Mediterranean region sandflies. The biological significance and pathogenicity of the virus carried by sandflies requires further investigation. In this study, the positive rates of Wuxiang virus neutralizing antibodies detected in the serum samples of local healthy people and domestic chickens were 8.4% (4/46) and 100% (4/4), respectively. Although the number of serum specimens collected in this study is limited, and only serum specimen from one animal (chicken)was collected, the above results suggest Wuxiang virus can infect humans and domestic chickens. Therefore, in the future, it is necessary to investigate the infection status of Wuxiang virus in local humans and animals to clarify the harm of the virus to public health.
The revised guidelines from ICTV meeting in 2020 recommended the addition of 53 new Phleboviruses (Kuhn et al. 2020). The previous ICTV classification guidelines included only 10 Phlebovirus species (Abudurexiti et al. 2019), including the SFTSV isolated in China (Yu et al. 2011) and the Hartland virus isolated in the USA (Shen et al. 2018). These two viruses are not classified as Phleboviruses according to the new ICTV guidelines, although 53 newly identified Phleboviruses were added. In this study, we conducted a molecular evolution analysis of the WUXV isolated in China in 2018 (Wang et al. 2020), and the newly identified Phleboviruses included in the revised ICTV classification guidelines. Among the new Phlebovirus species, 37 were isolated in the Americas (16 in North America and 21 in South America), 10 were isolated in Africa (1 in West Africa, 1 in East Africa, 3 in Central Africa, and 5 in North Africa), 8 were first isolated in Asia (1 in East Asia and 7 in West Asia), and 5 were from Europe (5 from Western Europe). A significant number of these Phleboviruses (n = 37 strains) uses sandflies as their transmission vector; 5 viruses are transmitted through mosquitoes, and one via ticks; the transmission vector is unknown for the remaining 17 viruses (Fig. 2). Here, we analyzed the M and S gene sequences of the four WUXV strains isolated from Yangquan sandflies (Fig. 2), and found that these isolates belonged to the same evolutionary branch as the TORV (Alkan et al. 2016); this suggested that WUXV may have derived from the TORV isolated from Turkey sandflies in 2012 (Alkan et al. 2016).
In this study, we re-isolated the WUXV from wild sandflies collected from Yangquan County, a different region from where the WUXV was originally isolated. These findings suggest that the WUXV has a relatively wide geographical distribution range. In addition, the neutralizing antibody of Wuxiang virus was detected in both human and chicken serum specimens (Table 5), suggesting that the local people and domestic chickens were infected with Wuxiang virus. Therefore, it is of great significance to strengthen the seroepidemiological investigation of Wuxiang virus in local humans and animals.
-
This work was supported by the Ministry of Science and Technology of the People's Republic of China (2018ZX10711001, 2018ZX10734-404-003);National Key Research and Development Program (No.2018YFA0900800); National Science and Technology Major Project (2018ZX1010 2001); National Natural Science Foundation of China (31900156); Key Research and Development (R & D) Projects of Shanxi Province, China (201803D31205); the United States National Institutes of Health U01 AI151810 and Development Grant of the State Key Laboratory of Infectious Disease Prevention and Control (2014SKLID103, 2015SKLID505).
-
QW and GL drafted and revised the manuscript. QW, SF, JC, XX, JW, BW, XT, YL, YH, FL, KN and SX participated in collection of sandfly specimens and acquisition of data. QW, SF, JC and GL participated in the analysis and interpretation of data. HW, XL and GL contributed to conception and design of the manuscript. All authors read and approved the final manuscript.
-
The authors declare no conflict of interests.
-
All animal experiments were conducted in strict compliance with the regulations set by the Animal Ethics Committee of China CDC.
















 DownLoad:
DownLoad: