HTML
-
Porcine epidemic diarrhea virus (PEDV) is an enteropathogenic alpha coronavirus which can cause porcine epidemic diarrhea (PED), a highly contagious swine disease that is prevalent among pig herds. PEDV was first identified in the 1970s, and in the following decades, it did not cause a significant impact on the pig industry until the emergence of new variant strains with high virulence in the 2010s. Since then, PEDV outbreaks have frequently occurred throughout the world, especially in China and some Asian countries (Lee 2015). Vaccination has been the primary measure for the prevention of PEDV. However, due to the emerging genetic variants, the current PEDV vaccines are not guaranteed to be effective. Generally, for the suckling piglets which are most vulnerable to PEDV, the mortality rate could reach 100% (Ma et al. 2015). Considering the great threat to the pig industry, it's meaningful to find more effective methods for the prophylaxis and treatment of PED in the long run.
Besides vaccination, some potential therapeutic methods for PED have also been reported, such as neutralizing antibodies (Gong et al. 2018; Zhang et al. 2019), anti-inflammatory factor (Li et al. 2019), peptides (Zhao et al. 2018), and small chemical molecules. Among them, small chemical molecules including natural products and synthetic compounds attract the most attention. Glycyrrhizin and glycyrrhizic acid, two active components of glycyrrhiza uralensis that is a common Chinese herb, have been characterized with various biological activities. Recently, the two compounds are found to effectively inhibit the PEDV infection in Vero cells by multisite inhibition mechanisms (Huan et al. 2017; Tong et al. 2020). The oleanane triterpenes and prenylated phenolic compounds from natural plants are also identified as PEDV proliferation inhibitors, some of them could inhibit viral replication at sub-micro molar level (Cho et al. 2019; Yang et al. 2015). In 2020, Peng et al. reported that GC376, a well-known broad-spectrum inhibitor of 3CLpro was highly effective against the PEDV 3CLpro with IC50 value of 1.11 µmol/L. The structural basis for inhibiting this enzyme was elucidated, providing important information for the structural optimization of GC376 (Ye et al. 2020).
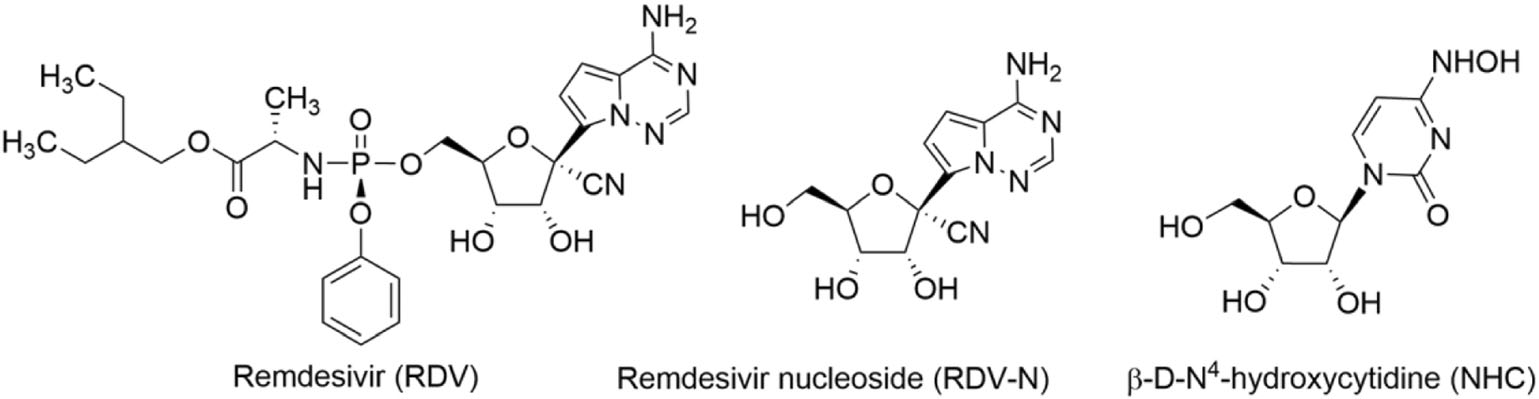
At the end of 2019, a new human coronavirus, named SARS-CoV-2 emerged and rapidly spread around the world, infecting nearly 98 million people and causing more than 2.0 million deaths by January 21st, 2021 (Valencia 2020). During this pandemic, an antiviral drug, remdesivir (RDV), which was previously indicated for Ebola virus infection, was authorized for emergency use against the viral infection on May 1st, 2020, and officially approved soon based on the positive results of three clinical trials (Beigel et al. 2020; Goldman et al. 2020; Spinner et al. 2020). RDV is a 1′-cyano-substituted adenosine nucleotide analogue prodrug which acts as a non-obligated chain terminator by targeting the highly conserved active site of viral polymerase and shows broad-spectrum antiviral activity against an array of RNA viruses, such as HCV, MERS virus, SARS-CoV virus, and Ebola virus (Agostini et al. 2018; Cho et al. 2012; Siegel et al. 2017). Moreover, this antiviral candidate is found to be able to inhibit zoonotic deltacoronaviruses with highly divergent RNA dependent RNA polymerases (RdRps), highlighting the value of this drug in the fighting against future emerging viruses (Brown et al. 2019). β-D-N4-hydroxycytidine, NHC, reported as early as 1959 (Fox et al. 1959) is also found to be a broad-spectrum antiviral nucleoside which has recently received a great deal of attention due to its potential for the treatment of SARS-CoV-2 infection (Sheahan et al. 2020b; Yoon et al. 2018).
As is well known, nucleoside analogs are an important source for discovering antiviral agents, and till now more than twenty-five drugs of this class have been approved for treating human viral diseases (Seley-Radtke and Yates 2018; Yates and Seley-Radtke 2019). However, with respect to animal viral diseases, little is known on the therapeutic potential of such kind of antivirals. PED is a serious disease that is lethal to piglet, and it is very meaningful to find effective anti-PEDV drugs. Herein, we evaluated the anti-PEDV activities of RDV, RDV nucleoside (RDV-N), and NHC, whose molecular structures were shown in Fig. 1. We found that RDV-N was the most active antiviral relative to its prodrug, RDV, and NHC. Moreover, RDV-N exhibited a high safety profile in mice which makes it to be a promising antiviral for the treatment of PEDV infection.
-
African green monkey kidney cells (Vero E6 cells) were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37 ℃ with 5% CO2. The PEDV variant strain SDSX16 (Accession no. MH117940.1) was isolated from a sucking piglet with acute diarrhea. Male ICR mice (18-22 g) were purchased from Shanghai Super-B & K Laboratory Animal Co. Ltd. (Shanghai, China).
-
RDV, RDV-N, and NHC were synthesized according to the reported methods (Purohit et al. 2012; Siegel et al. 2017) and the structures were determined by nuclear magnetic resonance. The samples used for in vitro and in vivo study were in a purity of at least 98.5% by high-performance liquid chromatography analysis.
-
To assess the cytotoxicity of RDV/RDV-N/NHC in Vero E6 cells, cells were seeded in 96-well plates and allowed to grow for 24 h to reach 80% confluence, and then treated with the test compounds (RDV/RDV-N/NHC) diluted with serum-free DMEM containing 10 μg/mL trypsin (Sigma, St. Louis, MO, USA) at a final concentration of 500 μmol/L, 250 μmol/L, 125 μmol/L, 62.5 μmol/L and 31.25 μmol/L. DMSO was used as the blank control. The cells were then cultured at 37 ℃ for 72 h, and the cytotoxicity was determined by CCK-8 assay.
-
Vero E6 cells were seeded in 96-well plates for 24 h and washed thrice with serum-free medium. PEDV (0.01 MOI) was first incubated with serial dilutions of the test compounds (RDV/RDV-N/NHC) at 37 ℃ for 1 h before infecting the Vero E6 cells. The final concentrations of RDV and RDV-N ranged from 125 μmol/L to 0.1 μmol/L, and the concentration of NHC ranged from 62.5 μmol/L to 0.5 μmol/L. DMSO was set as the blank control. The mixture was incubated at 37 ℃ until the cells of the virus control group showed complete cytopathic effect (CPE). The characteristic morphological changes of PEDV-infected cells in the antiviral groups were observed and photographed.
In another experiment, after adding the PEDV-compounds dilution to the cells, the mixture was incubated at 37 ℃ for 48 h and 72 h, respectively. Cells and supernatants were harvested to determine viral loads and titers using quantitative real-time RT-PCR (qRT-PCR) and 50% tissue culture infectious dose (TCID50) assay, respectively. The effective concentration that reached 50% decrease in viral replication was defined as the EC50 value.
-
Vero E6 cells were cultured in 24-well plates for 24 h and washed thrice with serum-free medium. PEDV (0.01 MOI) was first incubated for 1 h at 37 ℃ in the presence of 0.2 µmol/L RDV-N before infecting Vero cells. DMSO was used as the blank control. Viral propagation was confirmed by daily observation of the CPE. After 48 h at 37 ℃, cells were fixed with 4% paraformaldehyde for 15 min at room temperature (RT) and then blocked with 1% bovine serum albumin (BSA) in PBS for 30 min at RT. The monoclonal antibody specific against PEDV spike protein was added to the cells as primary antibodies and incubated for 1 h at 37 ℃. After washing three times with PBS, cells were incubated with FITC-conjugated anti-mouse IgG (Southern Biotech, Birmingham, AL, USA) in the dark for 30 min at 37 ℃. After washing five times with PBS, the images were captured using a fluorescence Leica DM IL LED microscope (Leica, Wetzlar, Germany).
-
96-Well plates of Vero E6 cells were prepared, infected, and treated similarly to those described above. After 48 h or 72 h, cells were harvested and total RNA was extracted from Vero E6 cells using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol, and was then reverse-transcribed into cDNA using oligo (dT) as the primer (Invitrogen). Relative quantitative real-time PCR was performed in a Roche Light Cycler 96 real-time PCR system. The real-time PCR results were analyzed and expressed as relative expression of CT (threshold cycle) valuing the 2−ΔΔCt method. The primers used for PEDV and GAPDH were listed below: PEDV-F: 5′-CGTACAGGTAAGTCAATTAC-3′; PEDV-R: 5′-GATGAAGCATTGACTGAA-3′; GAPDH-F: 5′-ACATCATCCCTGCCTCTACTG-3′; GAPDH-R: 5′-CCTGCTTCACCACCTTCTTG-3′.
-
Virus stock solutions described above (48 h post infection) were serially diluted before they were inoculated on the confluent Vero cell monolayers grown in the 96-well plates, followed by washing three times with PBS. Eight wells were inoculated with 100 μL at each dilution, and plates were incubated at 37 ℃ with 5% CO2 for 2 days. The wells with syncytium formation, the specific CPE caused by PEDV, were classified as PEDV-positive. PEDV titration was calculated by TCID50 following the Reed-Muench method.
-
Six male ICR mice were randomly divided into two groups. Group 1 was received a single intravenous (IV) administration of 25 mg/kg RDV-N dissolved in 5% DMSO, 5% ethanol, 40% PEG300, and 50% saline (pH 5-6 with 0.1 mol/L HCl), and Group 2 was received a single oral (PO) administration of 50 mg/kg RDV-N dissolved in DMSO/0.5% HPMC (5/95, v/v). The blood sample was collected at 5 min, 15 min, 30 min, 1.0 h, 2.0 h, 4.0 h, 8.0 h, and 24 h post-dosing, respectively, and the separated plasma was analyzed by LC/MS/MS. Mean plasma concentrations of RDV-N were used to calculate the pharmacokinetic parameters by noncompartmental analysis.
-
Fifteen male ICR mice were randomized into three treatment groups, with five animals per group. The three groups of animals were dosed once with RDV-N at 200, 500, and 1000 mg/kg orally, respectively (formulation: 5% DMSO + 5% Solutol HS15 + 90% saline). After administration, the mice were observed for 7 days (general signs, behavioral observation, body weight, and food consumption).
-
Vero E6 cells were seeded in a 6-well plate at 3.6 × 105 cells/well. After incubation for 24 h, half of the culture media (89% DMEM + 10% FBS + 1% sodium pyruvate) was removed from each well followed by the addition of 20 µmol/L RDV or RDV-N dissolved in the culture media and DMSO (99/1, v/v). The final concentration of RDV or RDV-N was 10 µmol/L (n = 3). Twenty-four hours later, the cells were harvested and lysed in 100 µL methanol/water (7/3, v/v). The nucleoside triphosphate metabolite was analyzed by LC/MS/MS.
Viruses, Cells and Mice
RDV/RDV-N/NHC
Cytotoxicity Assays
Anti-PEDV Activity Assay
Indirect Immuno-Fluorescence Assay (IFA)
PEDV Genomic and Subgenomic qRT-PCR Assay
TCID50 Assay
Pharmacokinetics Study of RDV-N in ICR Mice
Acute Toxicity Study of RDV-N in ICR Mice
Metabolism of RDV and GS-441524 in Vero E6 Cells
-
The cytotoxic effects of RDV, RDV-N, and NHC towards Vero E6 cell were determined before the antiviral assay. As shown in Fig. 2, both of RDV and RDV-N exhibited low cytotoxicity against Vero cell line with the maximal non-toxic concentrations (MNTC) of 125 µmol/L, and NHC showed a higher cytotoxicity with MNTC of 31.25 µmol/L.
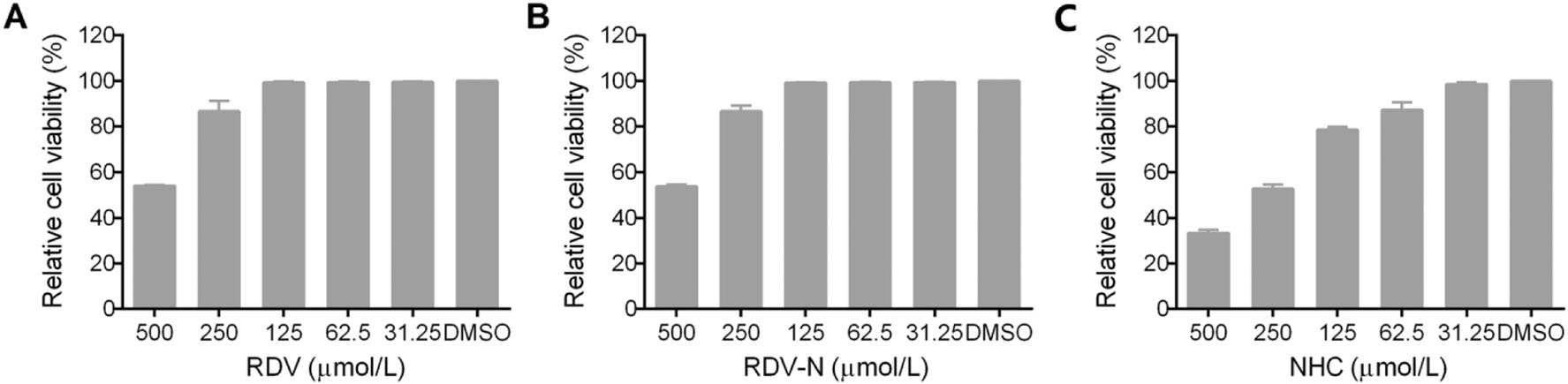
Figure 2. Cytotoxicity of RDV, RDV-N, and NHC in Vero E6 cells. Vero E6 cells were treated with indicated concentration of test compounds for 72 h, and cell viability was measured by CCK-8 assay. A The MNTC of RDV was 125 µmol/L. B The MNTC of RDV-N was 125 µmol/L. C The MNTC of NHC was 31.25 µmol/L.
-
PEDV infection of Vero cells can cause cytopathic effect (CPE). To determine whether the three nucleoside analogs possessed ant-PEDV activities, we investigated their effect on the morphological changes of PEDV-infected Vero E6 cells. As shown in Fig. 3, Vero E6 cells uninfected by PEDV were mainly in polygonal or irregular shape with clear outline (Fig. 3A), and not affected by 0.1% DMSO (Fig. 3B), while PEDV significantly caused cell fusion and the normal morphology of cells could not be observed (Fig. 3C). For the experiment groups, the PEDV-induced CPE was obviously blocked by the treatment with RDV, RDV-N, and NHC at the concentration of 0.5 μmol/L, 0.2 μmol/L, and 1.0 μmol/L, respectively (Fig. 3D-3F, Figures of high concentrations were not shown).
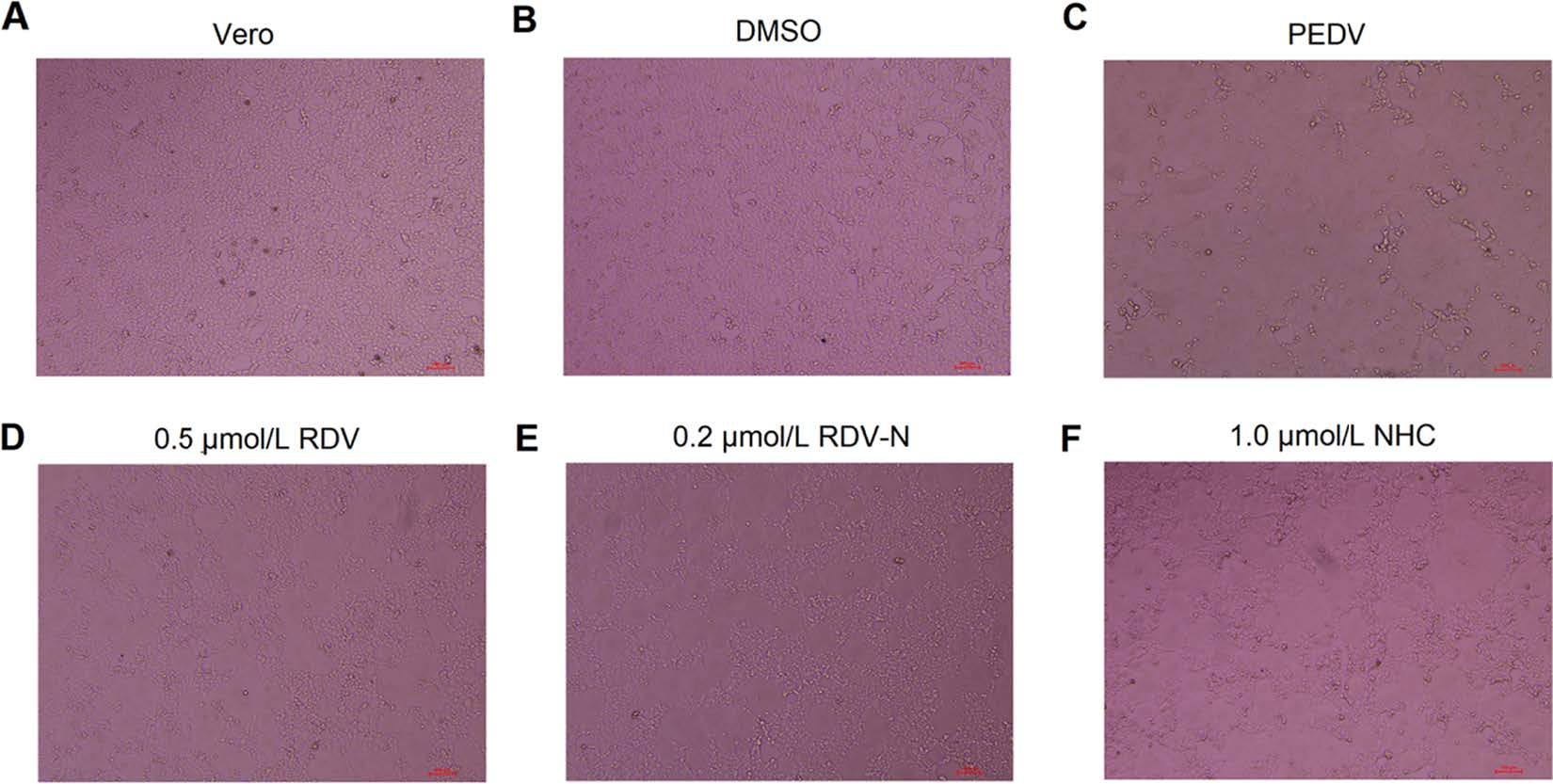
Figure 3. The effects of RDV, RDV-N, and NHC on PEDV-induced CPE. Vero E6 cells were seeded in 96-well plates for 24 h and washed thrice with serum-free medium. PEDV (0.01 MOI) was incubated with serial dilutions of test compounds at 37 ℃ for 1 h before infecting the Vero E6 cells. The mixture was incubated at 37 ℃ until the virus control group showed complete CPE. The characteristic morphological changes of PEDV-infected cells were photographed. A Non-infected cells. B Non-infected cells with 0.1% DMSO. C Cells infected with PEDV. D PEDV-infected cells with 0.5 μmol/L RDV. E PEDV-infected cells with 0.2 μmol/L RDV-N. F PEDV-infected cells with 1.0 μmol/L NHC (scale bar: 100 μm).
To measure the antiviral activities of the three compounds against PEDV, we used quantitative real-time RT-PCR (qRT-PCR) and TCID50 assay to determine the viral loads and titers in the infected cells, respectively. It was obvious to find that the viral genomic copies, as well as the viral titer, were decreased by all the three compounds in a dose-dependent manner at 48 h post infection (Fig. 4). Among them, RDV-N exhibited the strongest antiviral activity, and the virus replication was completely inhibited at the concentration of 2.0 μmol/L. Further, the EC50 values of them were determined at 72 h post infection using qRT-PCR. RDV-N had an EC50 value of 0.31 μmol/L which was twice active as its phospharamidate prodrug, RDV (EC50 = 0.74 μmol/L), and 3-folds more potent than NHC (EC50 = 1.17 μmol/L), shown in Fig. 5.
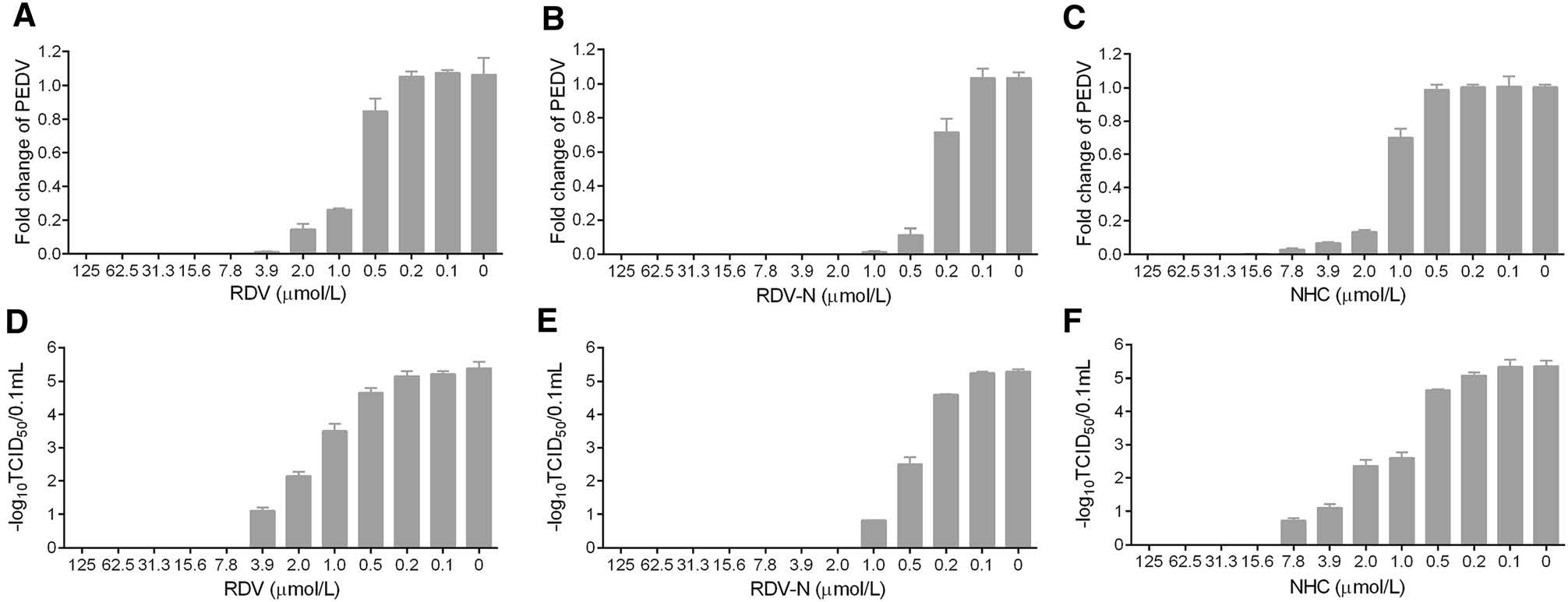
Figure 4. Reduction of PEDV replication by RDV, RDV-N, and NHC. PEDV (0.01 MOI) mixed with serial dilutions of test compounds was added to Vero E6 cells. After incubating at 37 ℃ for 48 h, cells and supernatants were harvested to determine viral loads (A-C) and titers (D-F) using qRT-PCR and TCID50 assay, respectively. A, D RDV completely inhibited PEDV replication at 7.8 μmol/L. B, E RDV-N completely inhibited PEDV replication at 2.0 μmol/L. C, F NHC completely inhibited PEDV replication at 15.6 μmol/L.

Figure 5. Dose-dependent curves of RDV, RDV-N, and NHC against PEDV. PEDV (0.01 MOI) mixed with serial dilutions of test compounds was added to Vero E6 cells. After incubating at 37 ℃ for 72 h, cells and supernatants were harvested to determine viral loads using qRT-PCR. The EC50 values were calculated from the dose response curve. A RDV had an EC50 of 0.74 μmol/L. B RDV-N had an EC50 of 0.31 μmol/L. C NHC had an EC50 of 1.17 μmol/L.
Moreover, we used an indirect immuno-fluorescence assay (IFA) to further confirm the anti-PEDV capacity of RDV-N. The viral protein expression level could be regarded as an indicator of viral quantity. From Fig. 6B and 6C, it can be found that the fluorescence intensity was significantly decreased in the presence of 0.2 μmol/L RDV-N, which strongly supported the potent antiviral activity of RDV-N.
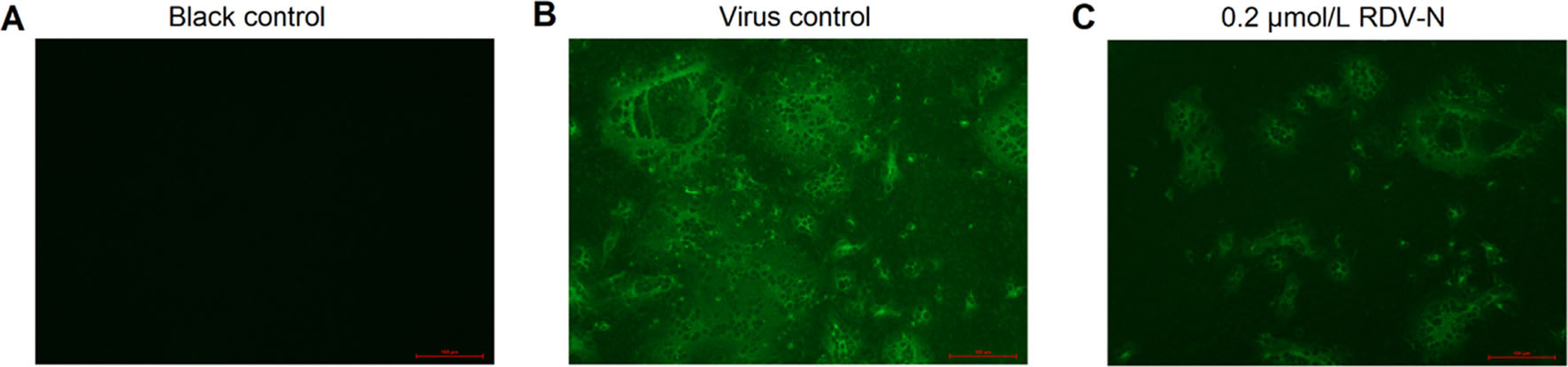
Figure 6. Immunofluorescent imaging of PEDV-infected Vero E6 cells. Vero E6 cells precultured in 24-well plates for 24 h were infected with PEDV (0.01 MOI) in the presence or absence of 0.2 µmol/L RDV-N. DMSO was used as the blank control. The mixture was incubated for 48 h followed by immunofluorescence imaging. A Blank control. B virus control. C 0.2 µmol/L RDV-N (scale bar: 100 μm).
-
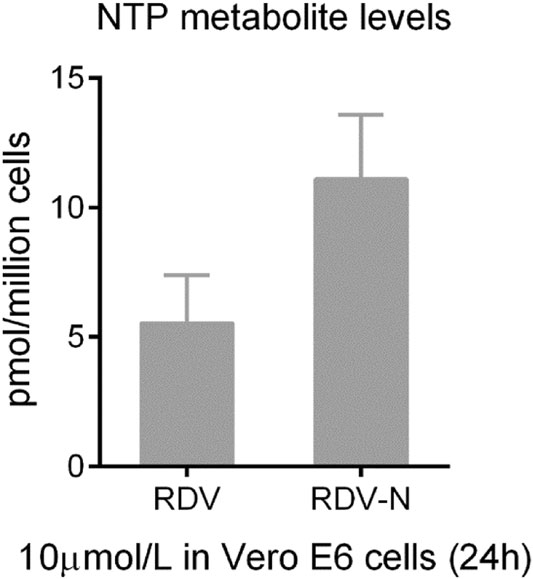
The pharmacokinetics of RDV-N was studied in ICR mice. As a nucleoside, RDV-N displayed poor water solubility and low lipophilicity. Predictably, this nucleoside demonstrated a low bioavailability (15.7%) in mice (Table 1). After oral administration, RDV-N rapidly reached the maximal plasma concentration within 1 h, and was eliminated quickly, similar to the data of intravenous administration. As an antiviral nucleoside, the active form of RDV-N was the nucleoside triphosphate (NTP) metabolite that mainly resided in tissues, and could hardly be detected in plasma, so we analyzed the in vitro metabolism of RDV-N and RDV in Vero E6 cells. As shown in Fig. 7, the two compounds were found to be metabolized to the corresponding triphosphate, and RDV-N gave a higher level than RDV (11.1 ± 2.5 vs 5.5 ± 1.8 pmol/million cells), which was consistent with the in vitro antiviral activities. For the acute toxicity study, the result showed that oral administration of RDV-N at the maximal dose of 1000 mg/kg did not cause animal death or marked toxic reactions.
Administration Dose (mg/kg) T1/2 (h) Tmax (h) Cmax (ng/mL) AUClast (h × ng/mL) F (%) PO 50 1.20 0.83 1826 4556 15.7 IV 25 1.02 – – 14,513 – Table 1. The pharmacokinetics of RDV-N in mice.
-
For RNA viruses, RdRp is the specific target of nucleoside antivirals, and the sequence similarity across different viruses is predictive for the antiviral spectrum of this kind of antivirals. Therefore, we performed an RdRp sequence similarity analysis among the six porcine coronaviruses (PEDV, PDCoV, TGEV, PRCV, PHEV, and SADS). The result indicated that PEDV (two strains: PEDV-CV777 and PEDV-SDSX16) shared a > 70% percent identity with SADS, TGEV, and PRCV, and a 60% percent identity with PHEV, but for porcine deltacoronavirus (PDCoV), the identity was only ~49%, shown in Fig. 8A.
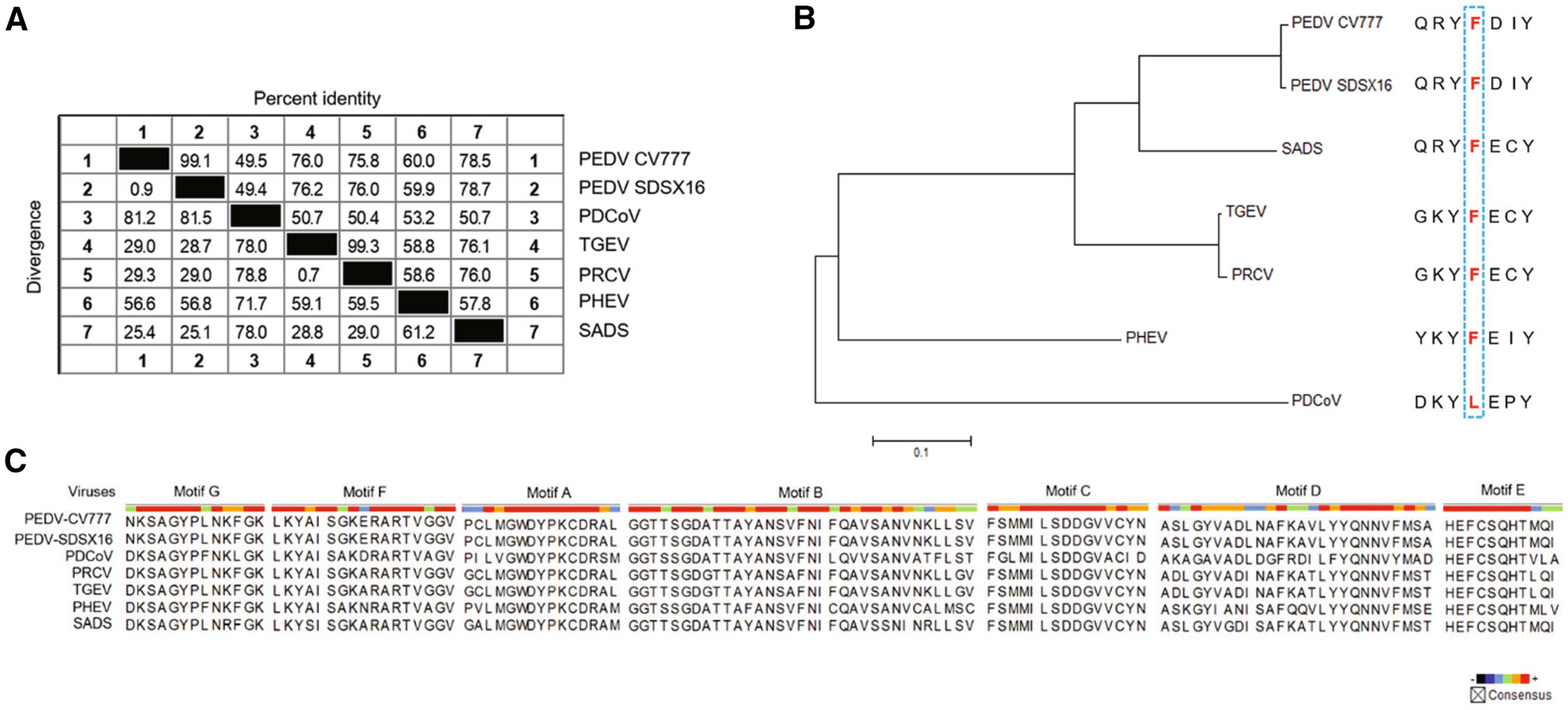
Figure 8. Similarity analysis in the RdRp of porcine coronaviruses. A Sequence similarity in the RdRp of the porcine coronaviruses (PEDV, PDCoV, TGEV, PRCV, PHEV, and SADS). PEDV shared a 49.4% to 78.7% percent identity with the other five viruses. B Genetic relatedness of the porcine coronaviruses RdRp proteins shown by phylogenetic analysis. PEDV was most similar to SADS, followed by TGEV/PRCV, PHEV and PDCoV. Among them, PDCoV harbored a natural Leu at the 483 position. C Amino acid multiple sequence alignments for each RdRp functional motif (A-G) of the porcine coronaviruses. The functional domain sequences of the two PEDV strains were 100% identical. The viruses, including PEDV, PRCV, TGECV, and SADS exhibited a high sequence similarity (> 90%) in Motif B and C.
It was reported that a Leu at 483 residue of deltacoronavirus RdRp, may be associated with partial resistance to RDV (Brown et al. 2019). Accordingly, we analyzed the 483 residue of the RdRps of the six porcine coronaviruses. It was found that only PDCoV harbored a Leu at the 483 position, where was a Phe for the other viruses (Fig. 8B). The coronavirus RdRps contain four highly conserved sequence motifs (A-D) that are involved in the catalytic process, and the functions of Motif B (substrate binding) and Motif C (positioning of the template-primer and facilitating the nucleotide incorporation) are well understood (Xu et al. 2003). We mapped the amino acid residues of these functional domains of the six porcine coronaviruses. The result indicated that the functional domain sequences (Motif A-G) of the two PEDV strains were 100% identical, and the viruses, including PRCV, TGEV, and SADS shared a high similarity (> 90%) in Motif B and C with PEDV (Fig. 8C).
Cytotoxic Activities of RDV, RDV-N, and NHC
Anti-PEDV Activities of RDV, RDV-N, and NHC
Pharmacokinetics, In Vitro Metabolism and Acute Toxicity Studies of RDV-N
Porcine Coronavirus Phylogenetic Analysis
-
As is known, antiviral nucleosides need to be converted to the triphosphate active forms to inhibit viral replication by acting as chain terminators or inducing viral lethal mutagenesis. During this process, the first phosphorylation step is regarded as the rate-limiting step (Jordheim et al. 2013). RDV is a phosphoramidate prodrug which can bypass the first step of phosphorylation. Such nucleoside prodrug form is characterized by many advantages (enhancing antiviral activity, improving pharmacokinetics properties, increasing therapeutic index, etc.) and has been widely applied in the antiviral drug discovery for hepatitis viruses (Mehellou et al. 2018). In our study, RDV exhibited lower anti-PEDV activity compared with its parent nucleoside (RDV-N), which was likely due to the deficiency of relevant esterases required to release the RDV nucleoside monophosphate in Vero E6 cells. In another aspect, it can be also speculated that the first phosphorylation step of RDV-N was not a major constraint to its antiviral effect. NHC, a nucleoside discovered very early has been extensively studied (Yoon et al. 2018). The 5′-isobutyrate ester prodrug of NHC, EIDD-2801 is now being evaluated for treating SARS-CoV-2 infection in Phase Ⅱ/Ⅲ clinical trials, and this drug shows favorable safety and pharmacokinetics properties based on the recently reported results of phase Ⅰ clinical studies (Painter et al. 2020). As a broad-spectrum antiviral nucleoside analog, EIDD-2801 is worthy of further studies in the treatment of other viral diseases.
RDV-N proved to be a very potent inhibitor of feline infectious peritonitis virus (FIPV), and could achieve a 100% cure rate in an experimental FIPV infection of cats by subcutaneous administration at a dose of 5 mg/kg or 2 mg/kg (Murphy et al. 2018). Our pharmacokinetics study in mice indicated that the oral bioavailability of RDV-N was low (15.7%), so this nucleoside is better to be administered by parenteral routes. Since mice have high levels of a serum esterase (carboxylesterase 1c, Ces1c) that is capable of hydrolyzing RDV, the pharmacokinetics study of RDV, as well as the in vivo antiviral assay was suggested to be performed in Ces1c−/− mice, and the relevant data are now available in several documents (Pruijssers et al. 2020; Sheahan et al. 2017, 2020a). Of note, RDV-N showed a good safety profile in cats, as evidenced by our acute toxicity study in mice. In a previous study, the cytotoxicity between RDV and RDV-N has been compared using a panel of human cell lines and primary cells. The result indicated that RDV with CC50 values in a range of 1.7-15.0 µmol/L was much more cytotoxic than RDV-N (CC50 > 100 µmol/L) (Warren et al. 2016). Indeed, the phosphoramidate ProTide approach is a liver-targeting technique because the esterases required to activate this prodrug are abundant in liver (Murakami et al. 2010). The clinical studies of RDV in patients revealed that the most common side effect was increased transaminases (Fan et al. 2020), which may be likely related to the properties of this prodrug. RDV-N was thought to have advantages over RDV for the treatment of COVID-19, and now it is being evaluated at the preclinical stage (Yan and Muller 2020). Accordingly, for PED that is an intestinal infectious disease, RDV-N may deserve further study to evaluate its therapeutic potential against this disease.
Besides PEDV, there are other coronaviruses which also can cause severe disease in the porcine herds (Wang et al. 2019). By analyzing the RdRp sequence similarity of six porcine coronaviruses, we found that PDCoV, an enteropathogenic coronavirus that causes serious vomiting and diarrhea in suckling piglets, exhibits a lower sequence similarity (~ 50%) among them. However, this virus is still susceptible to the antiviral activity (EC50 = 0.02 μmol/L) of RDV as reported in a recent research (Brown et al. 2019). As for the RdRp functional domains, which are responsible for binding with antiviral nucleotide and catalyzing the incorporation process, high sequence similarity was observed among PEDV, PRCV, TGEV, and SADS. Therefore, the three compounds, especially RDV and RDV-N are likely to be active against some of the other porcine coronaviruses.
In summary, we discovered that the broad-spectrum antiviral nucleoside analogs, RDV, RDV-N, and NHC were all potent PEDV replication inhibitors. Among them, RDV exhibited a promising prospect for the treatment of PEDV infection. It is also meaningful to investigate the antiviral potential of RDV-N against other porcine coronaviruses, and make some structural modifications on the scaffold to increase its oral bioavailability.
-
This work was supported by the following funding sources, a grant from the Shanghai Science and Technology Committee in China (Number: 19430750100), the National Science & Technology Major Project ''Key New Drug Creation and Manufacturing Program'', China (Number: 2018ZX09711002), the Shandong Key Provincial Research and Development Program (2019GNC106044) and the Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2016B14). We greatly appreciated Dr. Jia Liu and Wenjuan Hu for their help in the metabolism study of RDV and RDV-N.
-
BH and JS designed the experiments. YX, XG, TH, DW, XM and JW carried out the experiments. YX and XG analyzed the data, and wrote the paper. YX checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
All animal studies were conducted following protocols approved by the Institutional Animal Care and Usage Committee of Shanghai Institute of Materia Medica, Chinese Academy of Sciences.














 DownLoad:
DownLoad: