HTML
-
Since the first case of coronavirus disease 2019 (COVID-19) was reported as "pneumonia of unknown etiology" in early December 2019, this new coronavirus pneumonia epidemic caused by severe acute respiratory syndrome coronaviruses 2 (SARS-CoV-2), has spread around the world. As of Feb 24, 2021, there were 223 epidemic countries in the world, with 111,762,965 confirmed cases and 2,479,678 deaths (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The virus can infect people of almost all ages (Chen et al. 2020; Guan et al. 2020; Lu X et al. 2020). Although the mortality rate of SARS-CoV-2 is lower than that of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (2.2% vs 10% and 36%; De Wit et al. 2016), COVID-19 patients were often accompanied by multiple organ damage, including myocardial (Zeng et al. 2020), kidney (Diao et al. 2020), liver (Bangash et al. 2020; Zhao et al. 2020), male gonad (Ma et al. 2020), and even the central nervous system involvement, which may have lifetime impacts after recovery from infection. At present, a lot of issues needed to be addressed. For instance, the origin causing this outbreak are remaining unclear, although some related viruses were discovered on bats and pangolins (Lam et al. 2020; Zhang et al. 2020; Zhou et al. 2020). Global transmission pattern and genomic diversity are also essential to elucidate the dynamics for the pandemic.
Genome sequencing is an effective way to know the virus and thus to uncover its evolution. As exemplified in the Ebola outbreak in West Africa from 2013 to 2016, it was possible to reconstruct the spread, proliferation and decline of Ebola virus by analyzing the genome sequences of more than 5% of known cases (Dudas et al. 2017). At present, more genome sequences are still required, as about 0.54% genomes were deposited in public databases (include GISAID, GenBank, and NGDC). Therefore, an approach for rapid acquisition of SARS-CoV-2 genome is needed. Previous studies have shown that Oxford Nanopore sequencing possesses unique advantages in rapid accessing to pathogen genomes in infectious diseases like Lassa fever (Kafetzopoulou et al. 2019), Zika (Quick et al. 2017), Ebola (Quick et al. 2016), and ASFV (Jia et al. 2020) outbreak. Meanwhile, application of target-amplification would help to obtain high-quality virus genome from clinical samples (Ni et al. 2016; Chen et al. 2018; Faria et al. 2018). In this study, we further modified the approach that integrate SARS-CoV-2 target-amplification and Oxford Nanopore sequencing, and evaluate the approach by a set of clinical samples.
-
Respiratory specimens (swabs) collected from patients admitted to various Wuhan health care facilities were immediately placed into sterile tubes containing 3 mL of viral transport media (VTM). The swabs were deactivated by heating at 56 ℃ for 30 min in a biosafety level 2 (BSL 2) laboratory at the Wuhan Institute of Virology in Zhengdian Park with personal protection equipment for biosafety level 3 lab following the guidelines for detecting nucleic acid of COVID-19 in clinical samples. If the samples were not for use immediately, they were stored at 4 ℃.
-
Total nucleic acids were extracted using QIAamp@ 96 virus Qiacube HT kit on QIAxtractor Automated extraction (Qiagen, Hilden, Germany) following the manufacturer's instructions. A commercially acquired kit for SARS-CoV-2 Nucleic Acid Detection (Jienuo Company, Shanghai, China) was used. The kit is a one-step RT-qPCR kit designed to target the open reading frame 1ab (ORF1ab) and nucleocapsid protein (N) genes of the coronavirus. The N gene was labeled with a FAM reporter dye while the ORF1ab gene was labeled with a Texas red reported dye. The 20 μL reaction mixture consisted of 18 μL of freshly prepared mix and 2 μL RNA template. The one-step RT-qPCR protocol was run using the Bio-Rad's CFX 96 instrument under the following conditions: 42 ℃ for 5 min, 95 ℃ for 10 s, followed by 45 cycles of 95 ℃ for 10 s and reading at 60 ℃ for 45 s, respectively. Only positive or suspected positive samples will be sent for sequencing.
-
The nucleic acids were performed degeneration at 65 ℃ for 5 min, with two 23 μL volume reactions, including 5 μL Template RNA, 3.6 μL primer pool 1 or pool 2 and 13.65 μL nuclease-free water.
Then a one-step reverse transcription and amplicon amplification procedure was performed in each 50 μL total volume reaction, with 2× reaction mix (Thermo Fisher Scientific Inc., Massachusetts, USA), 2 μL SuperScriptTM Ⅲ RT/PlatinumTM Taq Mix and the previous 23 μL reaction solution. The PCR program settings are as follows: reverse transcription for 45 min at 50 ℃, thermal activation for 30 s at 98 ℃, followed by 35 cycles of denaturation at 98 ℃ for 15 s and annealing at 65 ℃ for 5 min, respectively.
The pool 1 and pool 2 amplification products of each sample were mixed and purified by the Agencourt AMPure XP beads at a 1:1 ratio, and finally diluted in 30 μL EB buffer. 1 μL purified DNA amplicons were used for quantification by Qubit (QubitTM dsDNA HS Assay Kit). The amplicons of each sample were then diluted to 1 ng/μL and 5 ng was used for Nanopore library construction. The sequencing library preparation consists of two steps: native barcode ligation and sequencing adapter ligation. The native barcoding of amplicons was performed in a 15 μL volume reaction (5 μL DNA amplicons, 7.5 μL nuclease-free water, 1.75 μL Ultra Ⅱ End Prep Reaction Buffer and 0.75 μL Ultra Ⅱ end Prep Enzyme Mix) for 10 min at room temperature and 5 min at 65 ℃ and 1 min on ice, then followed by a 50.5 μL total volume reaction (2.5 μL NBXX barcode, 17.5 μL Ultra Ⅱ Ligation Master mix, 0.5 μL Ligation Enhancer and previous 15 μL reaction solution) for 15 min at room temperature, 10 min at 70 ℃ and final 1 min on ice. Then mix all barcoded amplicons into one tube and quantify using Qubit. The sequencing adapter was ligated in a 50 μL volume reaction, with 30 μL barcoded amplicon pools, 5× NEBNext Quick Ligation Reaction Buffer, 5 μL AMⅡ adapter mix, and 5 μL Quick T4 DNA Ligase. The ligation reaction was performed at room temperature for 15 min. The library was purified using AMPure XP beads and quantified using Qubit.
Sequencing was performed on the PromethION platform, the final library was loaded onto the flow cell according to the manufacturer's instructions. ONT MinKNOW software was used to collect raw sequencing data, and Guppy was used for local basecalling of the raw data after sequencing runs were completed. Only reads with a mean quality score greater than seven were collected for subsequent analyses.
-
Firstly, sequencing data yielded from PromethION was filtered to remove low-quality reads with mean quality score less than seven. Secondly, data (reads quality score greater than seven) was subjected to remove reads shorter than 400 bp and longer than 600 bp by SeqKit (Shen et al. 2016). Thirdly, demultiplexing and adapters trimming were processed by qcat. The filtered data after using above three steps will be used as high-quality data for subsequent analysis.
-
The quality of sequencing data (high-quality data) for each sample was assessed by the following indicators: total bases, total reads number, reads length distribution, and mapped reads number, mapping rate of reads, coverage, mean sequencing depth, as well as median depth based on reference genome. Total bases, total reads number, and reads length were calculated by Perl language command. The mapping procedure was performed using Minimap2 (Li 2018) (with the parameters of -x map-ont) based on the genome sequence of IVDC-HB-01 (GISAID accession number: EPI_ISL_402119), and filtered by SAMtools (Li et al. 2009) (with the parameters of -F 3840 -q 60). Then the number of mapping reads, coverage length, and depth information of every site were obtained using the SAMtools (Li et al. 2009). Then the mapping rate of reads was calculated from the previous result of total reads number and mapping reads number, and the coverage rate of each sample was calculated from the coverage length information, and average depth and median depth were calculated from the depth information of every site using R language.
-
The correlation between Ct values and other different data quality indicators (total bases, mapping rate of reads, coverage, average depth, median depth) was examined by SPSS. First, tested whether each group of data obeys the normal distribution, and judged by skewness value, kurtosis value, significance value of Shapiro–Wilk test, and Q-Q plot. Secondly, the Pearson correlation coefficient test was carried out for the two sets of data that both obeyed the normal distribution, and the Spearman correlation coefficient test was performed on data that did not follow the normal distribution.
-
Mapping results were subjected to call SNPs using the tool Medaka (with the filter standards of ref_prob ≤ 0.01, QUAL ≥ 28, DP ≥ 15, AF ≥ 0.6 or ref_prob ≤ 0.06, QUAL ≥ 17, DP ≥ 30, AF ≥ 0.8) and the command bcftools mpileup (with the filter standards of depth of 10× and frequency of 0.6), followed by artificial verification, finally generated consensus using a script, margin_cons.py (Quick et al. 2017) (https://github.com/artic-network/fieldbioinformatics/blob/master/artic/margin_cons.py). Results of consensus sequences were used as genome sequences.
-
The reference genome sequence of IVDC-HB-01 (GISAID accession number: EPI_ISL_402119) and 38 genome sequences provided in the present study were aligned using MAFFT (Katoh et al. 2018), then used the alignment as input file of PERL script (available at https://github.com/zer0liu/bioutils/blob/master/snp/), which was developed to identify sites variations compared to the reference, at the same time, judge synonymous or non-synonymous mutations through the annotation of coding regions.
-
Amplicons of six selected samples (with more SNPs than the others) were directly ligated Illumina sequencing adapters using VAHTSTM Universal DNA Library Prep Kit for Illumina V3 (Vazyme Cat. ND607-01), then sequencing was conducted on the Miseq platform. The sequencing data were mapped to the reference genome of USA-CA1 (GenBank accession number: MN994467.1) with more variation sites compare to early Chinese isolate and performed SNP-calling by previously developed methods (Ni et al. 2016) (using standards of minor freq < 0.2 and major allele positive stripe to [0.0 to 1]). The differences in mutation sites and frequencies obtained by the two sequencing methods were compared for evaluation.
-
All the genome sequences of SARS-CoV-2 sequenced in the present study have been deposited in NGDC (accession no. GWHALPE01000000-GWHALPT01000000 and GWHALRI01000000-GWHALSH01000000) and GISAID (accession no. EPI_ISL_493149-EPI_ISL_493190).
Sample Collection and Preparation
Nucleic Acid Extraction and Viral Nucleic Acid Detection
Amplicon Nanopore Sequencing of SARS-CoV-2
Raw Data Processing
Sequencing Data Quality Assessment
Correlation Test Between Ct Value and Different Data Quality Indexes
Acquisition of SARS-CoV-2 Genome
Nucleotide and Amino Acid Substitution Recognition
Validation of Variation Sites and Mutated Allele Frequency between Oxford-Nanopore Sequencing and MiSeq Sequencing
Accession Codes
-
In order to rapidly obtain the genome sequence of SARS-CoV-2, the approach by utilizing SARS-CoV-2-specific amplicon amplification and Oxford Nanopore PromethION platform (https://artic.network/ncov-2019) was modified to reduce experiment time. This workflow started with the throat swab of the COVID-19 patient, which is widely used by all the countries, and followed by sample preparation, nucleotide extraction, cDNA synthesis and amplicon amplification in one step, and genome sequencing (Fig. 1). As viral load in swabs usually not abundant, we applied viral cDNA amplification before genome sequencing. According to the first released SARS-CoV-2 genome (Wuhan-Hu-1, GenBank accession: MN908947.3) (Wu et al. 2020), a set of 98 pairs of covering the nearly whole virus genome were used (https://github.com/artic-network/artic-ncov2019/tree/master/primer_schemes/nCoV-2019/V1). In general, the length of amplicons ranged from 381 to 420 bp, the melting temperature (Tm) is around 61 ℃, and the GC content (GC%) ranged from 30 to 54.55. The amplicons were designed overlapped, and covered 29,837 nucleotides, approximately 99.78% of the complete reference genome, including all coding regions and most of the UTR regions. Ideally, the entire process before sequencing would take 9 h and 20 min, and each person would be able to process 24 samples in a 96-well plate without affecting the overall process time. In the PromethION platform, one flow cell would yield about 2 Gb raw data per hour for the first 4 h, according to machine records. A total of 21 Gb raw data would yield after 12 h of sequencing. Eighteen to 24 samples would be sequenced simultaneously in a single flow cell. Hence, for a full day, more than 20 Gb raw data would be generated by a single flow cell, started from the throat-swabs. For a PromethION 48 sequencer, which can run up to 48 flow cells at once, 1152 samples can be sequenced in 12 h and yield exceed 1 Tb of raw data.
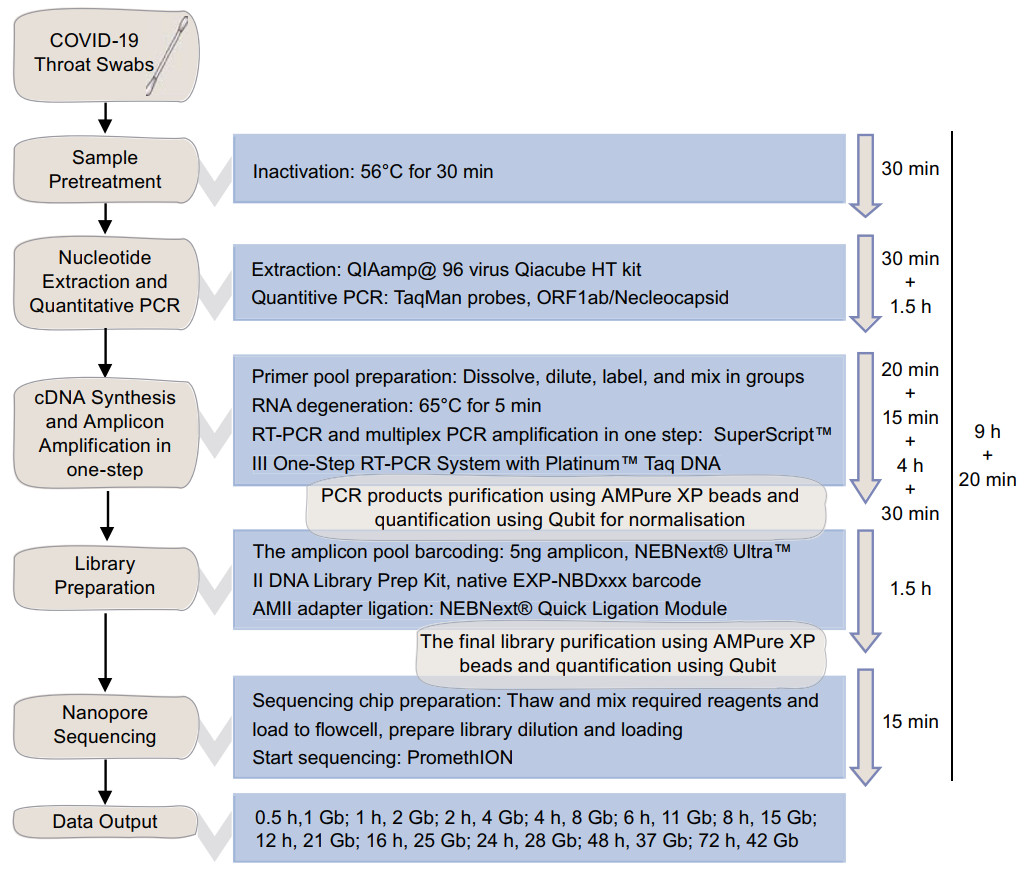
Figure 1. The workflow and schematic overview of SARS-CoV-2 genome sequencing using the amplicon-Nanopore technique. The general workflow of amplicon-nanopore sequencing of SARS-CoV-2 genome from throat swab samples.
As exemplified in this study, forty-two throat swabs from the COVID-19 patients were sequenced in two different batches of flow cells (Supplementary Table S1). The cycle threshold (Ct) values of those samples ranged from 18.74 to 39.14 (Supplementary Table S1). Following sequencing for about 71 h, a total of 85.74 Gb raw data were yielded. Firstly, 25% of low-quality reads (mean quality value less than seven) were filtered out, then 7% of reads with length greater than 600 bp and less than 400 bp from reads with mean quality score greater than seven were filtered out, followed by adapters and barcodes trimming, finally obtained 27.77 Gb high-quality data (the mean quality score is 9.89).
Then Minimap2 was applied for sequence mapping. After mapping to the reference genome of the first released virus isolate IVDC-HB-01 (GISAID accession number: EPI_ISL_402119), 38 samples (90.48%) with a depth greater than 100× with a genome coverage of about more than 90% were reserved for further analysis. For each sample, 196.71 Mb to 1,352.64 Mb (mean 723.64 Mb, median 697.82 Mb) high-quality data were kept, and the number of reads per sample ranged from 549,590 to 3,595,016 (mean 1,974,729, median 1,936,695). Of the total of 75,039,694 reads, 66,197,944 (88.22%) matched the reference genome, and for each sample, the mapped reads varied from 340,432 to 3,272,127 (mapping rate from 50.02% to 91.47%) (Fig. 2A, Supplementary Table S1). The high-quality read lengths ranged from 102 to 584 bp (mean 366 bp, median 386 bp) (Fig. 2B). The main peak of the length distribution was about 400 bp, which was consistent with the theoretical length distribution (peak at ~390 bp) of the amplicons (Fig. 2B). The over-length and under-length reads possibly originated from non-specific amplification in the multi-amplification step with so many primers, because the initial nucleic acid contains a large number of host and other microbial nucleic acids, therefore inevitably there will be interaction with homologous sequences of other species. Cause when divided the high-quality sequencing data into two parts with a threshold of 350 bp, classification of reads using Kraken2 (Wood and Salzberg 2014) showed that except for the major reads from SARS-CoV-2, there were reads from the cellular organisms and a small number of other microorganisms in both parts of data. In addition, the noticeable part of reads much shorter might be caused by partially degraded or mechanically sheared genome fragments, which may not completely cover every amplicon. As a result of when mapping to the SARS-CoV-2 reference genome showed that the two parts of the data were basically consistent with all data trends in mapping rate (Supplementary Fig. S1). In general, ~99.72% of the virus genome were mapped with reads, the mean depth ranged from 3996.3 to 39,045.6, and the median depth ranged from 441 to 35,678 (Supplementary Table S1). In Fig. 2C, we showed the mapping results of 10 representative samples with varied Ct values, from 18.74 to 39.14. As the Ct value increased, which means the viral load in the sample reduced, the quality of sequencing was decreased. The higher the Ct value, the more heterogeneous the mapping depth along the genome. For sample A164 (Ct = 18.74), there is only 0.19% genome with mapping depth < 100× and 1.83% genome with mapping depth < 1000×, whereas the sample C77 (Ct = 39.14), 10.63% of the genome had mapping depth < 100× and even 77.18% genome had mapping depth < 1000×.
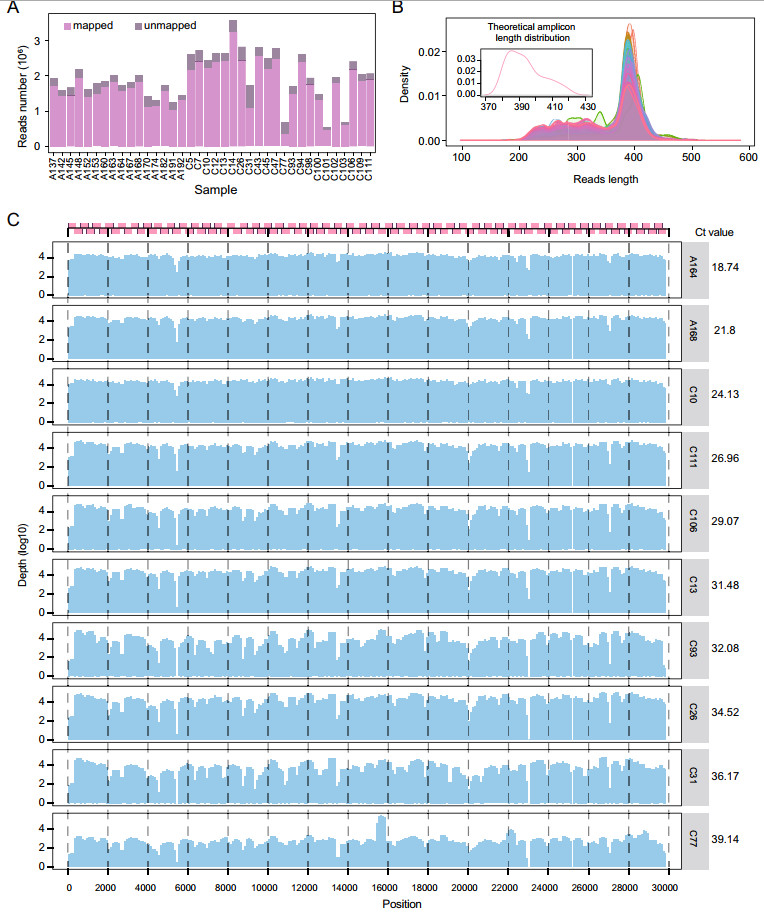
Figure 2. Performance of representative samples in the amplicon-Nanopore sequencing. A The proportion of mapped and unmapped reads of 38 samples when compared to the reference genome. Each bar represents a sample, bars in violet are reads mapped to the reference, and bars in dark-purple are unmapped reads. The order of samples is the same as the order of samples in Supplementary Table S1, excluding 4 low-quality samples. B The length distribution of reads. The internal small graph represents the reads distribution according to the theoretical length of amplicons, while the external large graph represents the reads length distribution of each sample in the actual sequencing results. C Sequencing coverage and depth overview of 10 samples with different Ct values. On the left is the corresponding situation of amplicons location and sequencing coverage and depth of each sample genome, and on the right is the Ct value of each sample.
-
The Ct value and different sequencing quality indicators were further assessed, and found that the total output high-quality bases, mapping rate, the coverage of genome, and the mean and median of mapping depth all were negatively correlated to the Ct value (Fig. 3A, 3B), though three indicators (total high-quality bases, average depth, and median depth) were not affected significantly (P-value > 0.05) in the sequencing batch PAE38111 (Fig. 3B). Moreover, data yielding for low and high Ct values were assessed. For sample A164 (Ct = 18.74), reads saturation reached after 8 h of sequencing, 99.84% of the genome had mapping depth > 100×, and 98.29% genome had mapping depth > 1000× (Fig. 3C). For samples with Ct value between 20 and 25 (n = 8), sequencing saturation almost reached after 10 h of sequencing, 95.75% of genome had mapping depth > 100× and 82.98% of genome had mapping depth > 1000× (Fig. 3D). For samples with Ct value between 25 and 30 (n = 12), sequencing saturation reached nearly after 12 h of sequencing, 93.82% of genome had mapping depth > 100× and 81.36% of genome had mapping depth > 1000× (Fig. 3E). For samples with Ct value between 30 and 35 (n = 14), sequencing saturation almost reached after 16 h of sequencing, 89.42% of genome had mapping depth > 100× and 73.95% of genome had mapping depth > 1000× (Fig. 3F). For samples with Ct value greater than 35 (n = 3), sequencing saturation almost reached after 16 h of sequencing, 85.37% of genome had mapping depth > 100×, and 52.11% of genome had mapping depth > 1000× (Fig. 3G). Of course, the longer the sequencing time, the better the mapping results. The saturation value may increase again for longer sequencing as showed in Fig. 3E–3G, but it is not necessary for urgent requirements. Sequencing time can also be adjusted specifically based on real-time analysis of Oxford Nanopore sequencing.
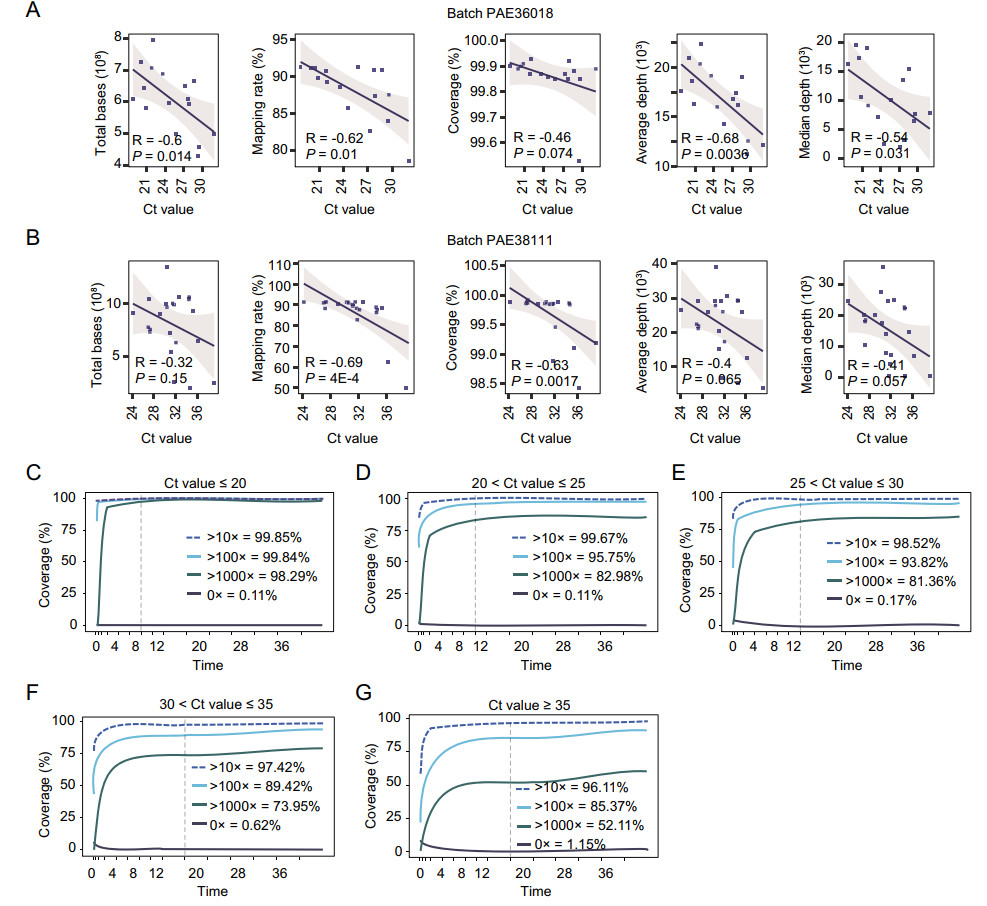
Figure 3. The correlation between different data quality indicators and Ct value and data yielding assessment. A, B Correlation between Ct value and five sequencing quality indicators (total data volume, mapping rate, coverage, average sequencing depth, median sequencing depth) in the sequencing batch PAE36018 (A) and batch PAE38111 (B). C–F The genome coverage process with sequencing of samples in different range of Ct values: C, Ct values less than 20 cycles (n = 1); D, Ct values between 20 and 25 (n = 8); E, Ct values between 25 and 30 (n = 12); F, Ct values between 30 and 35 (n = 14); G, Ct values greater than 35 (n = 3). The coverage of the data when mapped to the reference genome at different sequencing time points: the black line represents the proportion of unmeasured regions to the genome, the blue dotted line represents the proportion of regions with sequencing depth greater than 10, the light-blue line represents the proportion of regions with sequencing depth greater than 100, and the green line represents the proportion of regions with sequencing depth greater than 1000. The gray vertical dotted line indicates the time at which the sequencing basically reached saturation. The proportion indicated in the figure is the data of sequencing time point of corresponding saturation timepoint.
-
By using bcftools mpileup and Medaka for SNP calling and a published script (margin_cons.py; Quick et al. 2017) for consensus generation, 38 nearly full-length (on average about 99.61%) SARS-CoV-2 genomes with high-quality, and 4 shorter genomes (from 93.92 to 97.68%) with gaps were obtained. All these genomes have been deposited in NGDC (accessions: GWHALPE01000000-GWHALPT01000000 and GWHALRI01000000-GWHALSH01000000) and GISAID database (accessions: EPI_ISL_493149-EPI_ISL_493190). Compared to an early virus isolate genome IVDC-HB-01 (GISAID accession number: EPI_ISL_402119), 39 SNP sites were discovered in the 38 nearly full-length genomes, with 18 synonymous mutation sites and 21 nonsynonymous ones (Fig. 4, Supplementary Table S2). For each virus genome, there were 0 to 5 SNPs compared to IVDC-HB-01. For all SNP sites, only one mutated nucleotide existed. Through comparison with all deposited genome sequences in GISAID database, as of submitted date of March 1,2020,7 of the SNPs were appeared in previous released genome sequences (before the date of the given sample collected) as well as observed in the following virus genomes, 32 SNPs were firstly discovered at the date of sample collection (Supplementary Fig. S2, Supplementary Table S2).

Figure 4. Nucleotide and amino acid variations in 38 sample genomes. The top shows the open reading frames (ORFs) position of the reference genome, and the middle displays the amino acid variations on the genome of each sample. The blue triangle represents synonymous mutation, the red triangle represents non-synonymous mutation. The bottom shows the nucleotide composition at the mutation sites identified in the 38 samples. Different colors represent different bases, the bar in red is adenine (A), the bar in green is cytosine (C), the bar in blue is thymine (T), and the bar in purple is guanine (G).
Due to the existence of obvious part of reads shorter than the theoretical amplicon length, whether these abnormal reads had an impact on the accuracy of the genome needs to be addressed. On the one hand, the results mapping to the SARS-CoV-2 reference genome showed that the sum of the coverage of the genome (Supplementary Fig. S1B) and the average sequencing depth (Supplementary Fig. S1C) obtained from the separate two-part (> 350 bp or < 350 bp) data were basically the result from the total reads. On the other hand, reads shorter than 350 bp account for about 29% (13.4%–62.6%) of all reads, and contributed about 21% (9.5%–54.9%) of the genome sequencing depth, therefore when removed data shorter than 350 bp, there was but only a very slight effect on the SNP presence in the genome: in 42 samples, only two SNPs were inconsistent with that from total data: the mutated allele frequency (MuAF) at 20,170 (A170, 0.7 vs. 0.54) and 10,894 (A192, 0.71 vs. 0.53) were slightly higher than that in the total data.
-
Given that Oxford Nanopore sequencing had a higher error rate for single read (Jain et al. 2016), whether deep sequencing and bioinformatics would eliminate sequencing error is of concerns. To address this issue, six samples (A191, C10, C14, C31, C43, C106) were subjected to Illumina MiSeq platform, since they had more SNPs than the others. The same amplicon amplification products were used for Illumina library preparation. From the MiSeq platform, a total of 5.6 Gb of raw data were generated (11,711,358 paired-end reads). Genome coverage ranged from 98.81% to 99.90%, and 89.24% to 99.78% of the genome regions had sequencing depths higher than 10 × accordingly. Through comparison of results from Oxford Nanopore PromethION and Illumina MiSeq platforms, we found that 45 out of 50 SNPs discovered by Nanopore sequencing can also be found by Illumina sequencing, while 2 SNPs discovered by Illumina sequencing were failed to uncover by Nanopore sequencing (Fig. 5, Supplementary Table S3). For the six samples, the average error rate was less than 0.4 per 10, 000 bp. Of note, the false positive and false negative SNPs only existed in two samples (A191 and C31), in which the concentration of purified amplicons was lower than 1 ng/μL. Thus, the quality of the sequencing library may affect the sequencing results to a certain extent.
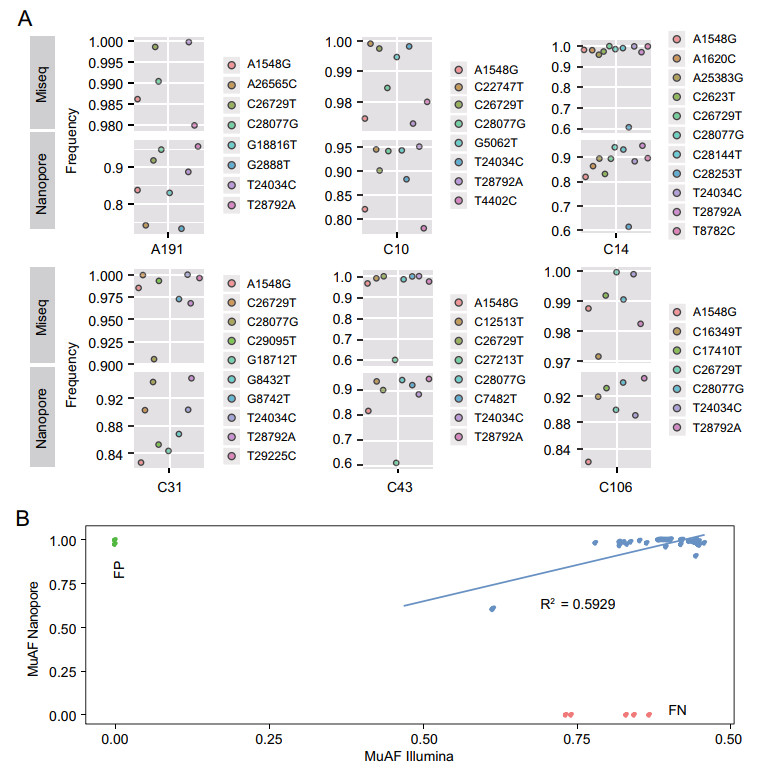
Figure 5. Evaluation of variation sites and mutated allele frequency of Nanopore sequencing using Miseq sequencing. A Comparison of mutation frequency of SNP sites obtained by Miseq sequencing analysis with those obtained by Nanopore sequencing analysis in six samples with more SNP sites than the others. The ordinate is the proportion of the number of major mutant bases to the total number of all sequenced bases at the mutation site. The various mutation types are shown in different colors and are marked on the right side of each small figure. B Correlation of mutated allele frequencies (MuAFs) observed for SNPs detected at viral genomes with Nanopore and Illumina sequencing. SNPs detected with Nanopore but not Illumina were considered to be false-positives (FP; green) and SNPs detected with Illumina but not Nanopore were considered to be false-negatives (FN; red).
Moreover, MuAFs of each variation sites were further compared. For this, SNP-calling pipeline were conducted for determination of variation sites. The results showed that the proportion of the dominant variant type calculated by Nanopore sequencing was significantly lower than that of Illumina sequencing in 42 out of 45 simultaneous sites (t-test, P < 0.05, Fig. 5, Supplementary Table S3). The MuAFs of these sites ranged from 0.02 to 0.2 less on the Nanopore platform than on the MiSeq platform, with an average of 0.09 less per site. And the MuAFs of the three exceptional sites were about 0.02 more (no significance, t-test, P = 0.183) on the Nanopore platform than on the MiSeq platform. This possibly reflected the difference of sequencing and bioinformatic procedures between Nanopore and Illumina platforms.
Amplicon Sequencing of SARS-CoV-2 from COVID-19 Swab Samples Based on Oxford Nanopore
The Quality of SARS-CoV-2 Genome Sequencing is related to the Viral Load of the Sample
High-quality Genome Revealed Genetic Variations of SARS-CoV-2
NGS Sequencing Verifies the High Accuracy of Amplicon Sequencing of SARS-CoV-2 Based on Oxford Nanopore
-
When dealing with the emerging infectious diseases, acquiring the genome of the causative pathogens is a top priority in early anti-epidemic works, as virus genomics is one of the direct ways to understand the etiology of an emerging infectious disease. Moreover, virus genome information can help researchers to carry out pathogen identification, important genes analysis, origins tracing, dynamic tracking and transmission and epidemic prejudgment etc. For the COVID-19 epidemic caused by SARS-CoV-2, direct detection of clinical samples is the fastest way to obtain the viral genome, such steps like cell culture (Kim et al. 2020), virus isolation (Park et al. 2020) can be omitted. The most commonly used method was the metagenomic next-generation sequencing (mNGS) (Zhou et al. 2020), it has unique advantages in the screening of unknown pathogens. But for known pathogens, mNGS will need more data than amplicon sequencing for genome acquisition because clinical samples such as that of COVID-19 patients (most are oropharyngeal swabs) often have low viral nucleic acid load and complicated background, which increases the sequencing cost and analysis cost to some extent. On the other hand, because complete genome coverage cannot be guaranteed, Sanger sequencing is often used to fill in the gaps, which increases workload and time cost.
With the continuous development of sequencing technology, Oxford-Nanopore sequencing has become one of the powerful means for the rapid detection of pathogens. Its rapid, portable, and real-time characteristics make it played an important role in the outbreak of Lassa fever (Kafetzopoulou et al. 2019), Zika (Quick et al. 2017), Ebola (Quick et al. 2016), and other infectious diseases. In the current COVID-19 pandemic, some research teams have also improved their SARS-CoV-2 whole-genome sequencing (WGS) methods in other ways, such as increasing the length of amplicons to reduce costs (Freed et al. 2020), combining multi-target amplification and rapid barcode library preparation to shorten time costs (James et al. 2020), and using transposase mediated addition of adapters and PCR based addition of symmetric barcodes to increase throughput (Baker et al. 2020). In the present study, we merged the reverse transcription (RT) PCR and amplicons amplification into one-step to shorten experiment time, and comprehensively evaluated this type of amplicon-Nanopore sequencing technology.
First of all, results demonstrated that high-quality SARS-CoV-2 virus genome covering all ORF regions could be obtained from clinical samples within 24 h (Figs. 1, 3). Since the coverage and depth of the viral genome were evaluated with the change of sequencing time, recommended sequencing time was given for samples with different Ct value ranges (Fig. 3C–3G), which can effectively guide the reasonable sequencing arrangement. Moreover, the throughput of Nanopore sequencers can meet the needs of large-scale sequencing, and a single Nanopore PromethION 48 sequencer can process more than 1000 samples per day. According to our rough estimate, the total cost of nucleic acid positive detection and sequencing for a sample is less than $170, which is equivalent to the generally accepted low-cost mNGS sequencing. In addition, there is still potential for further improvement in genome collection as we used the primers V1 published by the ARTIC network, and the ARTIC team is constantly optimizing the primer pool. Only in the current version, the Ct values of the clinical pharyngeal swab samples we evaluated ranged from 18.74 to 39.14 (Supplementary Table S1). Compared with other studies using Nanopore sequencing (Baker et al. 2020; Lu J et al. 2020), 42 samples sequenced in this study showed better results, 38 samples (90.5%) covered more than about 90% of the genome with sequencing depth of more than 100×, and the other 4 samples (9.5%) covered more than 70% of the genome with sequencing depth over 100×. Furthermore, Illumina sequencing verified that the high error rate for single read in Nanopore sequencing (Jain et al. 2016) could be reduced or even be completely eliminated via ultra-high deep sequencing in the Nanopore sequencing platform: in the six verification samples, the overall error rate is less than 0.4 per 10, 000 bp; At the same time, 2/3 of the samples are 100% accurate (Fig. 5), and through the backtracking of the experimental process, it was found that 1/3 of the samples with false positives or false negatives may be caused by the low sample quality. Since the current SARS-CoV-2 virus genome variations are mostly random mutations (Supplementary Fig. S1), systematic errors in the genome sequences obtained by Nanopore sequencing is completely negligible in large-scale genomic analysis.
However, since the frequency of major mutant alleles in Nanopore sequencing is significantly lower than that in Illumina sequencing (Fig. 5, Supplementary Table S3), that is, there are still many minor mutations caused by technical errors in nanopore sequencing, which is the same as previous studies (Harel et al. 2019), so other auxiliary methods are needed in the analysis of intra-host mutations (e.g., iSNV).
-
The study was supported by grants from the Foundation for National Mega Project on Major Infectious Disease Prevention (grant number 2017ZX10103005-005), National Key Research and Development Program of China (2020YFC0845800 and 2020YFC0845600), and the National Natural Science Foundation of China (31970548 and 91631110). We thank the ARTIC-network for publishing their amplicon primers, we thank Lei Zhang, Ding Gao, Juan Min, Anna Du, Dongbo Nie of the core facility and technical support at Wuhan Institute of Virology, as well as Tao Du of National Biosafety Laboratory, Wuhan, Chinese Academy of Sciences for assistance with experimental platform and experimental environment maintenance.
-
This project was designed by Jianjun Chen, DL, HW. Samples were collected and prepared by Jianjun Chen, Jun Chen, YH, YZ, JX. Experiments were conducted by Jianjun Chen, Jun Chen, YY, KW, WQ, YL. The methods were developed by Jianjun Chen, DL, HW, YY, HL, XW, KH, ZJ, DW. The data analysis was performed by YY, KW, HL, XW. The manuscript was prepared by Jianjun Chen, DL, HW, YY. All authors read and commented on the paper.
-
All authors declare that they have no conflict of interest.
-
The study and use of all samples were approved by the Ethics Committee of Wuhan Pulmonary Hospital (No. 2020-LS-001), consents from patients were waived by the Ethics committee.














 DownLoad:
DownLoad: