HTML
-
Rotavirus (RV) is a major pathogen leading to the acute viral gastroenteritis in young children and animals. RV genome contains 11 double stranded RNA fragments, encoding 12 proteins, including 6 structural and 6 non-structural proteins (Estes 2013). Based on VP6, RVs can be divided into ten different species by now, including A-H, one tentative species Ⅰ, and a candidate species J (Banyai et al. 2017). The RV particle consists of three capsid layers. Two major surface proteins, VP7 and VP4, formed the outer most layer and played an important role in inducing neutralizing antibodies and protective immunity. VP7 and VP4 defined the RV G and P genotypes, respectively (Matthijnssens et al. 2011). The group A RVs are diverse and has been identified 51 P genotypes so far (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/7th-RCWG-meeting). VP4 can be cleaved by trypsin into two fragments, VP5* and VP8* (Fiore et al.. 1991). VP8* located at the distal part of the VP4 spike and was reported to be essential in the glycan recognition and cell attachment (Dormitzer et al. 2002a).
RVs were identified in many species, including human, porcine, bovine, dog and birds. Bats, the reservoirs of many viruses, were also identified to harbor the RVs (He et al. 2013). The genetic analysis of bat RVs indicated the bat-to-human transmission and reassortment (He et al. 2017; Komoto et al. 2020). The cross-species transmission of RVs has been reported in several RV genotypes, including P[6], P[3] RVs that were reported in both human and animal infections (Nyaga et al. 2018; Okitsu et al. 2018). P[3] RVs were identified in various species, including human, simian, dog, and bat (Tsugawa and Hoshino 2008; He et al. 2013; Sasaki et al. 2016). Bat P[3] LZHP2 was identified in an insectivorous bat and shared high nucleotide identity with the unique human strain M2-102 that was suggested to originate from a bat RV as a result of cross-species transmission and was divergent from other human RV strains (Dong et al. 2016; He et al. 2017). Human HCR3A was supposed to be of dog origin (Tsugawa and Hoshino 2008). The molecular mechanism of the prevalence of these P[3] RVs remained elusive.
Attachment to the cell receptors is crucial for the virus infection and transmission. The illustration of the interactions between virus and ligand provided the basis for understanding host tropism, host adaptation, and zoonosis. Previous studies have shown that some animal RVs recognize sialic acid-containing glycoconjugates and are neuraminidase sensitive (Ciarlet and Estes 1999; Isa et al. 2006). Some animal RV VP8*s, including P[1] bovine Nebraska calf diarrhea virus, P[2] simian SA11, P[7] porcine CRW-8 were reported to interact with terminal sialic acids on the cell surface glycans (Ciarlet and Estes 1999). The simian P[3] Rhesus rotavirus (RRV) VP8* recognized N-acetylneuraminic acid (Neu5Ac) and the crystal structure of RRV VP8* in complex with Neu5Ac has also been determined (Dormitzer et al. 2002b). The glycan binding site located at one corner of the two beta-sheets. CRW-8 P[7] VP8* was found to bind the sialic acid using the same glycan binding site as that of RRV P[3] (Blanchard et al. 2007).
Though the interaction between RRV P[3] and sialic acid has been reported, the glycan binding specificities of the P[3] RVs of other species is not clear yet, especially the bat P[3] RVs. Here, the functional features of bat, human, and dog P[3] RVs were investigated, which deepened the understanding of P[3] RV prevalence and transmission from animals to human.
-
The full VP8* fragments (residues 1–231 amino acids) of human HCR3A (Genebank: EU708904), 12,638 (Genebank: LC340022), M2-102 (Genebank: KU597745), dog RV198-95 (Genebank: HQ661137), and bat LZHP2 (Genebank: KX814943) were synthesized by Genewiz company (Suzhou, China) and cloned into pGEX6P-1 with an N-terminal GST tag. The GST-fusion proteins were expressed in E.coli and induced with isopropyl-β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mmol/L at 22 ℃ for 16 h. The recombinant proteins were purified as previously reported (Ma et al. 2015; Sun et al. 2018). Briefly, the bacteria were collected and resuspended, followed by ultrasonication. The supernatant of the bacterial lysates was filtered through a 0.22 μm membrane (Millipore) and then loaded onto the glutathione sepharose (GE Healthcare Life Sciences). The column was washed with PBS five times. Then the protein of interest was eluted with buffer of 50 mmol/L Tris–HCl, 10 mmol/L reduced glutathione, pH 8.0. The eluted proteins were concentrated and the buffer was changed to PBS. The protein concentration was determined using the BCA kit (BD Biosciences). Bat P[3] mutant of C189Y and human M2-102/HCR3A mutant of Y189C were cloned using the site-directed point mutation. The mutant proteins were expressed and purified as described above. The eluted proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
-
Enzyme-linked immunosorbent assay (ELISA)-based oligosaccharide binding assays were conducted as previously described (Sun et al. 2020). The GST-VP8* fusion proteins were coated onto microtiter plates at 20 μg per well with incubation at 4 ℃ overnight. Following blockage with 5% nonfat milk, synthetic oligosaccharides conjugated with polyacrylamide (PAA) and biotin were added at 0.2 μg per well. The following oligosaccharides were used in this study: Neu5Ac, Neu5Gc, Neu5Acα2-3Gal, Neu5Acα2-6Gal, siaLea, siaLex, siaLec, Neu5Acα2-3Galβ1-4GlcNAc (3'SLN), A/B disaccharides, H1, H2, lea, lex, leb, ley, mucin core 1–mucin core 6, and core 8 (GlycoTech, USA). Then, horseradish peroxidase (HRP)-conjugated streptavidin (Abcam) was added at 0.1 μg per well. For each step, the plates were incubated at 37 ℃ for 1 h, and washed five times with 0.5% PBS-Tween 20 buffer between steps. The color reactions were developed using a 3, 3', 5, 5'-tetramethylbenzidine (TMB) kit (BD Biosciences). The absorbance of 450 nm was measured.
-
Glycan ligand screening for human M2-102 and bat LZHP2 P[3] VP8*-GST fusion proteins were performed by the Protein-Glycan Interaction Core of the Consortium for Functional Glycomics (CFG) (http://www.functionalglycomics.org/) against a library containing 600 glycans. The identities of all glycans in Version 5.3 of the array are available at http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml. The recombinant GST-VP8* protein was used at protein concentrations of 5 μg/mL and 50 μg/mL, respectively. The bound GST-VP8* proteins were detected using a fluorescence-labeled anti-GST monoclonal antibody (Sigma). Fluorescence was measured and quantified by a microarray scanner and relative fluorescent unit (RFU) for binding to each glycan was calculated (Cholleti et al. 2012).
-
The interactions between P[3] VP8*s and different oligosaccharides were further analyzed using Octet RED96 (ForteBio., USA). Oligosaccharides with PAA-biotin tag (10 μg/mL) (Neu5Ac, Neu5Gc, Neu5Acα2-3Galβ1-4GlcNA(3'SLN), Neu5Acα2-6Gal, H1, H2, H3, Neu5Acβ2-6(Galβ1-3)GalNAc, B disaccharide, A disaccharide, B tetrasaccharide, core1, core2, core8, siaLea, siaLex, siaLec, lea, leb, lex, ley) were immobilized on the streptavidin-biosensor. The P[3] VP8* proteins (human HCR3A, M2-102, bat LZHP2) were diluted at 10 μmol/L with PBST buffer (PBS with 0.02% Tween-20, 0.5% BSA) and added to the 96-well microplate. The VP8* proteins were injected and binding responses were measured. The apparent equilibrium dissociation constants for VP8* and oligosaccharides were calculated using Octet analyzing software.
-
The GST-VP8* proteins were twofold serially diluted with a starting concentration of 2 mg/mL using PBS buffer and added to 96-well V-bottom plates (Costar) with 50 μL per well. Animal red blood cells (RBCs) (Gelaidisi, Beijing) of porcine, rabbit, bovine, chicken, lamb, and human RBCs (A, B, O types, donated by lab colleagues) were diluted to 1% with PBS. Written informed consents were signed by the colleagues. Authorization for the sampling of human and animal blood was obtained. Then 50 μL of RBC was added to each well. Agglutination was determined after incubation at 25℃ for 1 h.
-
A homology model of bat P[3] VP8* was constructed based on the structure of RRV P[3]VP8* (PDB identifier [ID] 1KQR) by SWISS-MODEL automated protein structure homology modeling server (http://swissmodel.expasy.org/). The structural superimposition and analysis were performed using pymol (https://pymol.org/2/). The cartoon and surface models of VP8* were shown. The electrostatic potential presentation was calculated with the pymol software.
Protein Expression and Purification
Oligosaccharide Binding Assay
Glycan Microarray Screening Analysis
Biolayer Interometry (BLI) Analysis
Hemagglutination
Homology Modeling and Structural Analysis
-
P[3] VP8*s of human (HCR3A, 12,638, M2-102), dog (RV198-95), and bat (LZHP2) were expressed in a soluble form with an N-terminal GST tag. The GST-VP8* proteins were purified via affinity chromatography and the molecular weight were about 52 kDa with free GST protein of 26 kDa (Fig. 1).
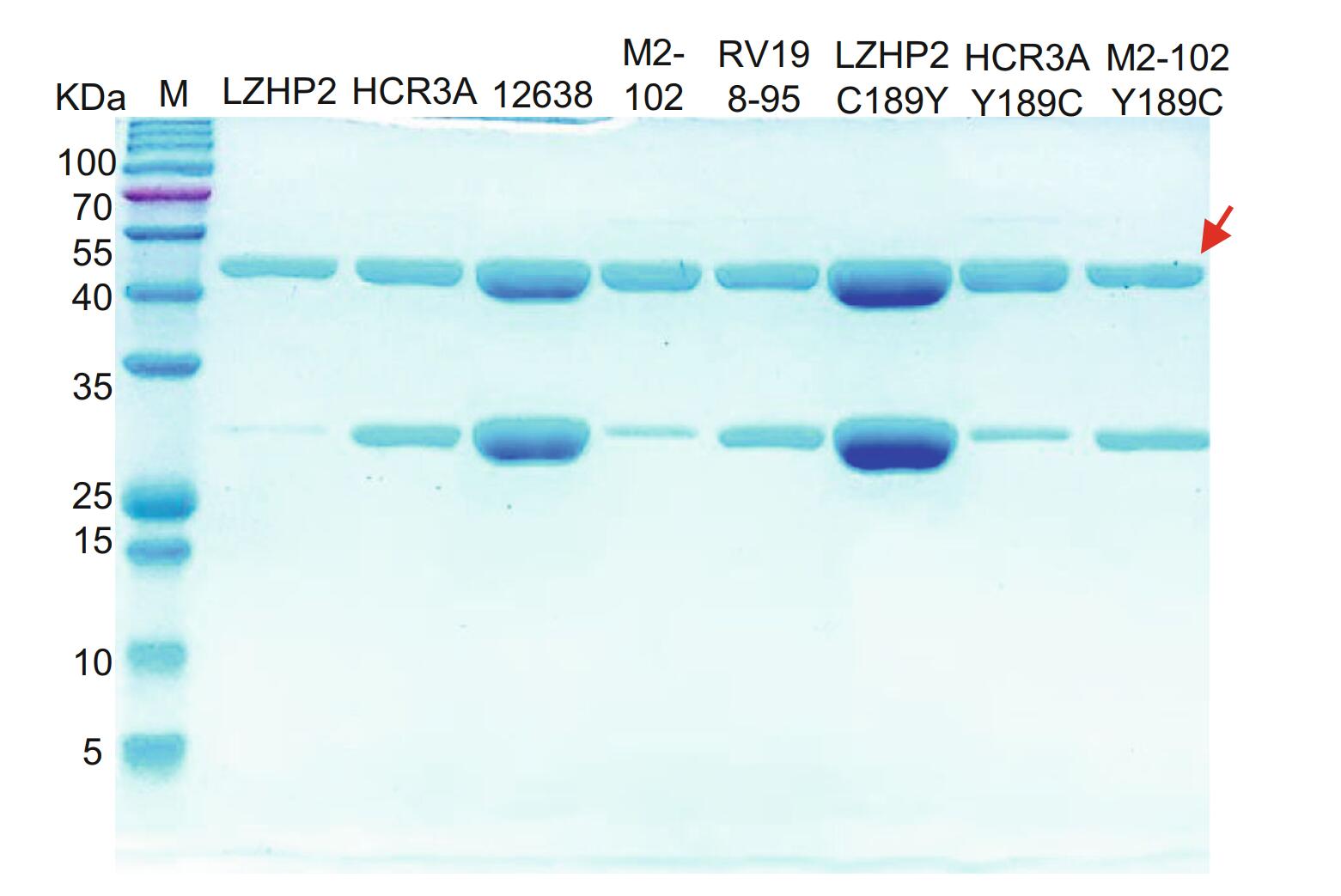
Figure 1. SDS-PAGE of bat LZHP2 P[3], human HCR3A, 12,638, M2-102 P[3], dog RV198-95 P[3], bat P[3] LZHP2 C189Y, human HCR3A Y189C, human M2-102 Y189C VP8* proteins. The molecular weight of GST-VP8* protein is ~ 52 kDa. The 26 kDa band indicates the free GST protein. The red arrow refers to the protein of interest.
-
VP8* proteins were applied to the ELISA to investigate the binding to synthetic oligosaccharides. Human HCR3A, 12,638, and dog RV198-95 P[3] VP8*s exhibited binding to glycans with sialic acid, such as Neu5Acα2-3Gal, Neu5Acα2-6Gal, and 3'SLN (Fig. 2). Human M2-102 and bat LZHP2 P[3] VP8*s showed no observed binding to the tested glycans.
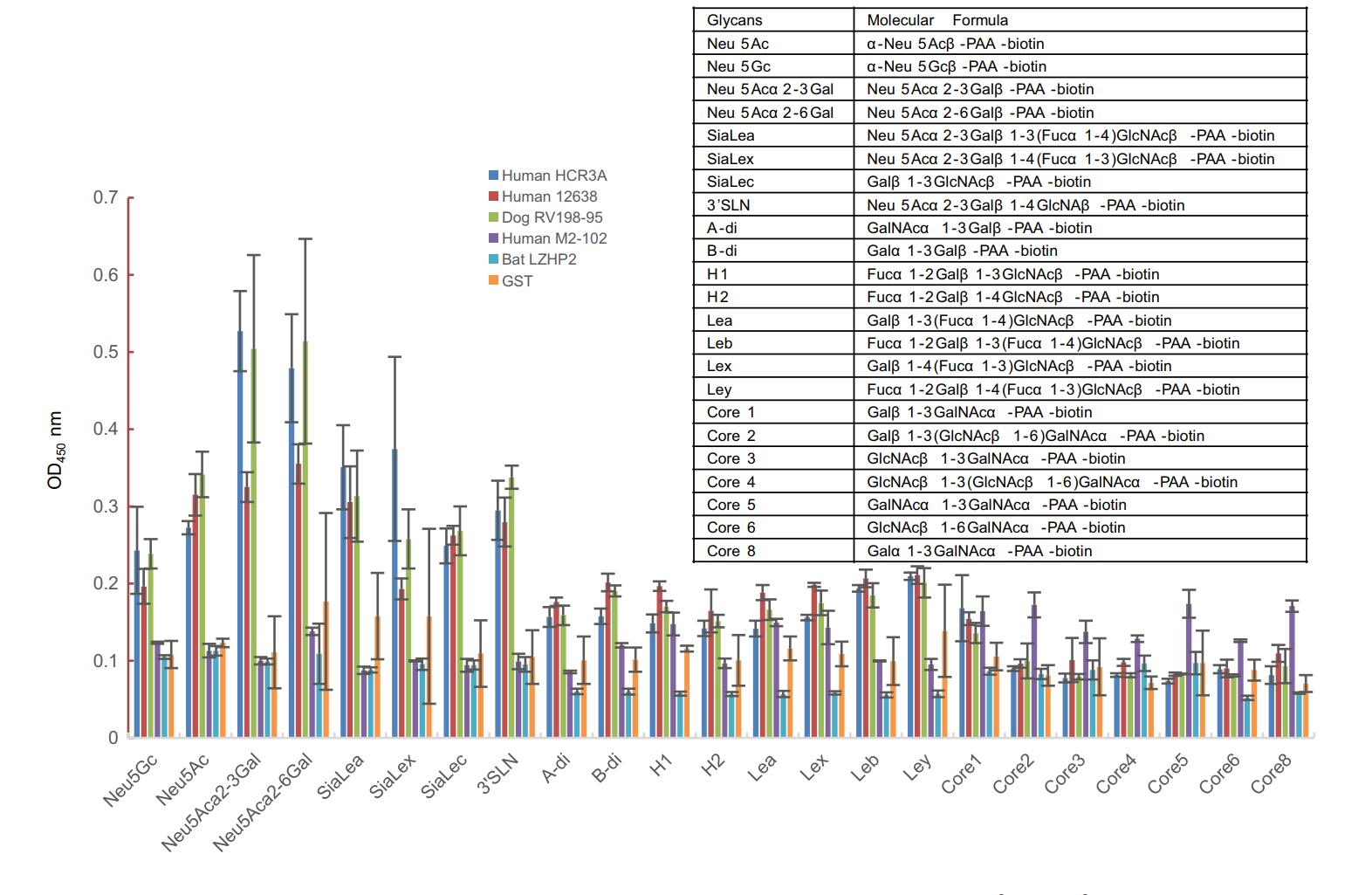
Figure 2. The glycan binding specificities of P[3] RV VP8*s. PAA-biotin oligosaccharides such as Neu5Ac, Neu5Gc, Neu5Acα2-3Gal, Neu5Acα2-6Gal, siaLea, siaLex, siaLec, Neu5Acα2-3Galβ1-4GlcNAc (3'SLN), Neu5Acβ2-6(Galβ1-3)GalNAc were included. The absorption at OD450 nm was detected. The molecular formula of these glycans were listed in the table.
According to the glycan microarray assay, human M2-102 and bat LZHP2 P[3] VP8* exhibited no specific binding to the tested 600 glycans including glycans with sialic acid (Fig. 3). Relative Fluorescent Units (RFUs) indicated the binding intensity. The Human M2-102 and bat LZHP2 P[3] VP8* did not showed high RFU value to these glycans (Fig. 3). For the glycan 117, the RFUs were almost the same for different concentrations and different kinds of proteins. It is suggested that the binding is regarded to be non-specific.
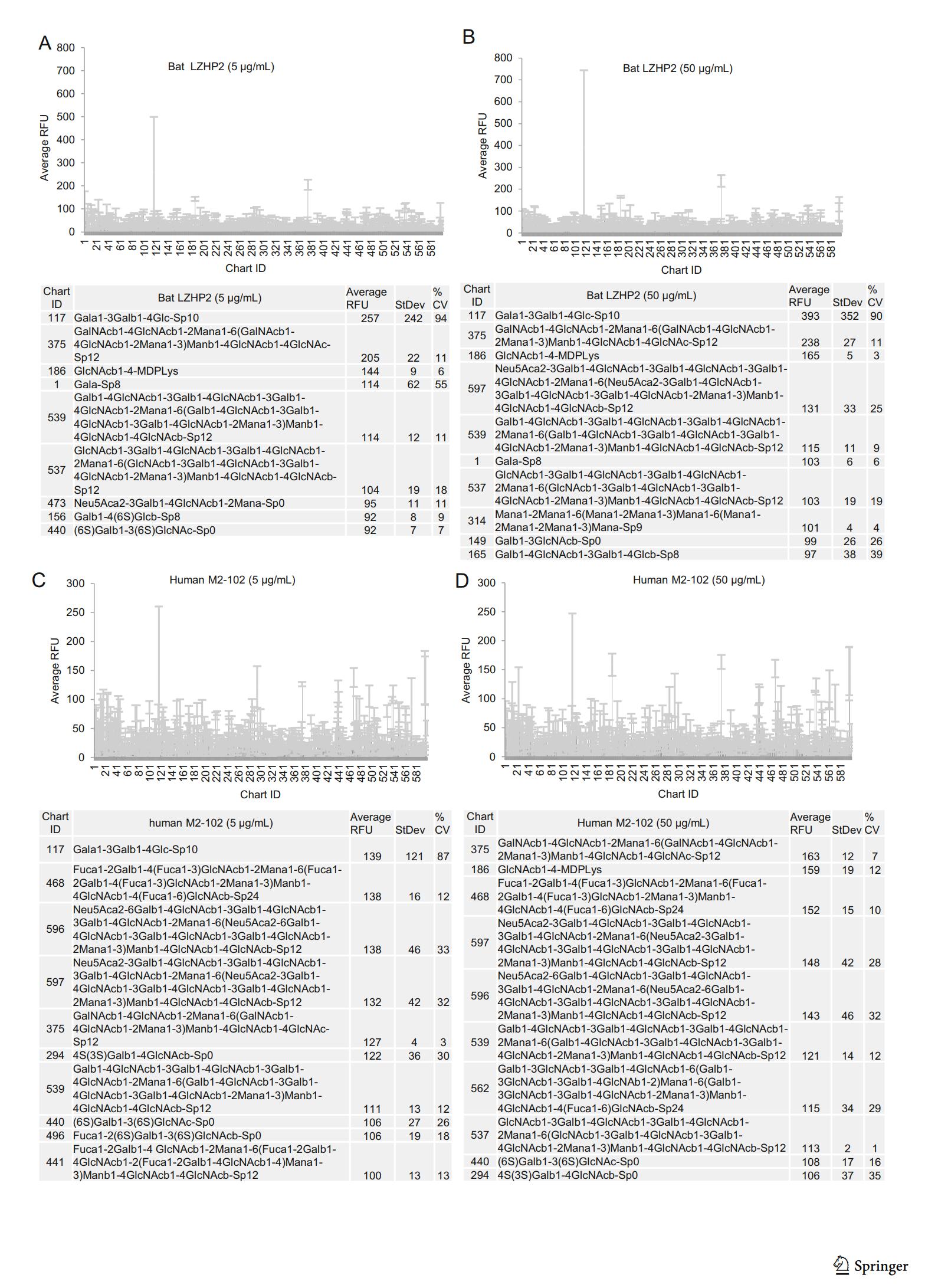
Figure 3. Graphical representation of P[3] VP8* (bat LZHP2 (up panel), human M2-102 (bottom panel)) VP8* at (A/C) 5 μg/mL, and (B/D) 50 μg/mL binding to the array of 600 glycans. Relative Fluorescent Units (RFU) correspond to the strength of binding to individual glycans. Binding specificity is shown in mean relative fluorescence units (RFU) of binding to n = 6 replicates of each glycan printed on the array and % coefficient of variation (%CV). The highest and lowest point from each set of 6 replicates has been removed so the average is of 4 values. The strength of binding at two concentrations is shown. Data for the top 10 of the glycans bound with highest intensity are listed in each table.
To test the interactions between P[3] VP8*s and different glycans, the biolayer interferometry (BLI) assay was performed using the Octet RED96. The results showed that human HCR3A VP8* specifically recognized glycans with sialic acid, including Neu5Ac, 3'SLN, Neu5Acα2-6Gal, siaLec, siaLea, siaLex (Fig. 4), while P[3] VP8*s of human M2-102 and bat LZHP2 did not recognize any tested glycans.

Figure 4. Biolayer interometry (BLI) analysis. PAA-biotin labeled oligosaccharides, Neu5Ac, Neu5Gc, Neu5Acα2-3Galβ1-4GlcNA-(3'SLN), Neu5Acα2-6Gal, H1, H2, H3, Neu5Acβ2-6(Galβ1-3)GalNAc, B disaccharide, A disaccharide, B tetrasaccharide, core1, core2, core8, siaLea, siaLex, siaLec, lea, leb, lex, ley were loaded on the biosensor at 10 μg/mL. Human HCR3A, M2-102, and bat LZHP2 P[3] VP8* proteins were diluted at 10 μmol/L and applied to the association and dissociation procedures. The glycan binding data was analyzed by the Octet analyzing software.
-
Hemagglutination was performed to explore the interaction between VP8* and the red blood cells. Human HCR3A, 12,638, M2-102, and dog RV198-95 P[3] VP8*s hemagglutinated the red blood cells of porcine, chicken, rabbit, bovine, lamb, human A, B, O types (Fig. 5). But bat LZHP2 P[3] VP8*s did not hemagglutinate any cells.

Figure 5. The hemagglutination assay of P[3] RV VP8*s. Red blood cells of chicken, porcine, rabbit, bovine, human A, B, O were used in the assay. The protein at 2 mg/mL was diluted serially with PBS as shown. PBS buffer was used as the negative control (NC).
It is noted that the VP8*s with Y189 could hemagglutinate the red blood cells. In order to test the function of residue 189, we constructed the mutants of bat LZHP2 P[3] C189Y, human HCR3A Y189C and M2-102 Y189C. Bat P[3] C189Y could hemagglutinate the red blood cells, whereas human HCR3A P[3] Y189C and M2-102 Y189C showed no hemagglutination (Fig. 5).
-
Sequence alignment showed that bat LZHP2 P[3] VP8*s possessed various amino acids with the sequence identity of 84.4%, 84.4%, 97.4%, 84.0% compared to VP8*s of human HCR3A, 12,638, M2-102, dog RV198-95 P[3], respectively. The residues involved in the glycan binding were diverse, especially residue 189 that was essential for the sialic acid binding (Dormitzer et al. 2002b). Bat LZHP2 P[3] VP8* possessed 189 cysteine (C189), while all the other tested P[3] VP8*s had tyrosine (Y189) (Fig. 6). V144 and T191 in bat LZHP2 P[3] VP8* are the same as that of bat-origin human M2-102, while human HCR3A, 12,638, and dog RV 198–95 P[3] VP8*s possessed S144 and A191. Residues R101, T146, Y155, G187, Y188, S190 are relatively conserved in all these P[3] VP8*s except H155 in human HCR3A.

Figure 6. VP8* protein sequences of different P[3] strains were aligned using DNAMAN. The residues involved in the ligand binding were colored red. The amino acids that are different between bat and human/dog were presented in green. H155 in human HCR3A VP8* was colored blue. Residue C189 was colored yellow. GeneBank numbers of the sequences are: bat LZHP2 (KX814943), human M2-102 (KU597745), human HCR3A (EU708904), human 12,638 (LC340022), dog RV198-95 (HQ661137).
-
The homology model of bat P[3] VP8* was constructed based on the P[3] RRV crystal structure (PDB ID: 1KQR) by SWISS-MODEL. The structural superimposition showed that bat P[3] VP8* model presented typical galectin-like conformation, but it is quite different to RRV P[3] VP8* with the root mean square deviations (RMSD) values of 1.256 (Fig. 7A). Y189 in RRV P[3] formed hydrogen bonds with sialic acid, while C189 would lost the interaction (Fig. 7A), which may affect the ligand recognition. V144 and T191 may influence the interactions via the loop conformation, which still need further study to clarify. The surface presentation exhibited that the putative sialic acid binding site was discontinuous in the P[3] bat VP8* model (Fig. 7B). The electrostatic potential was also different with positive charge in P[3] bat VP8* model and negative charge in P[3] RRV VP8* (Fig. 7C).
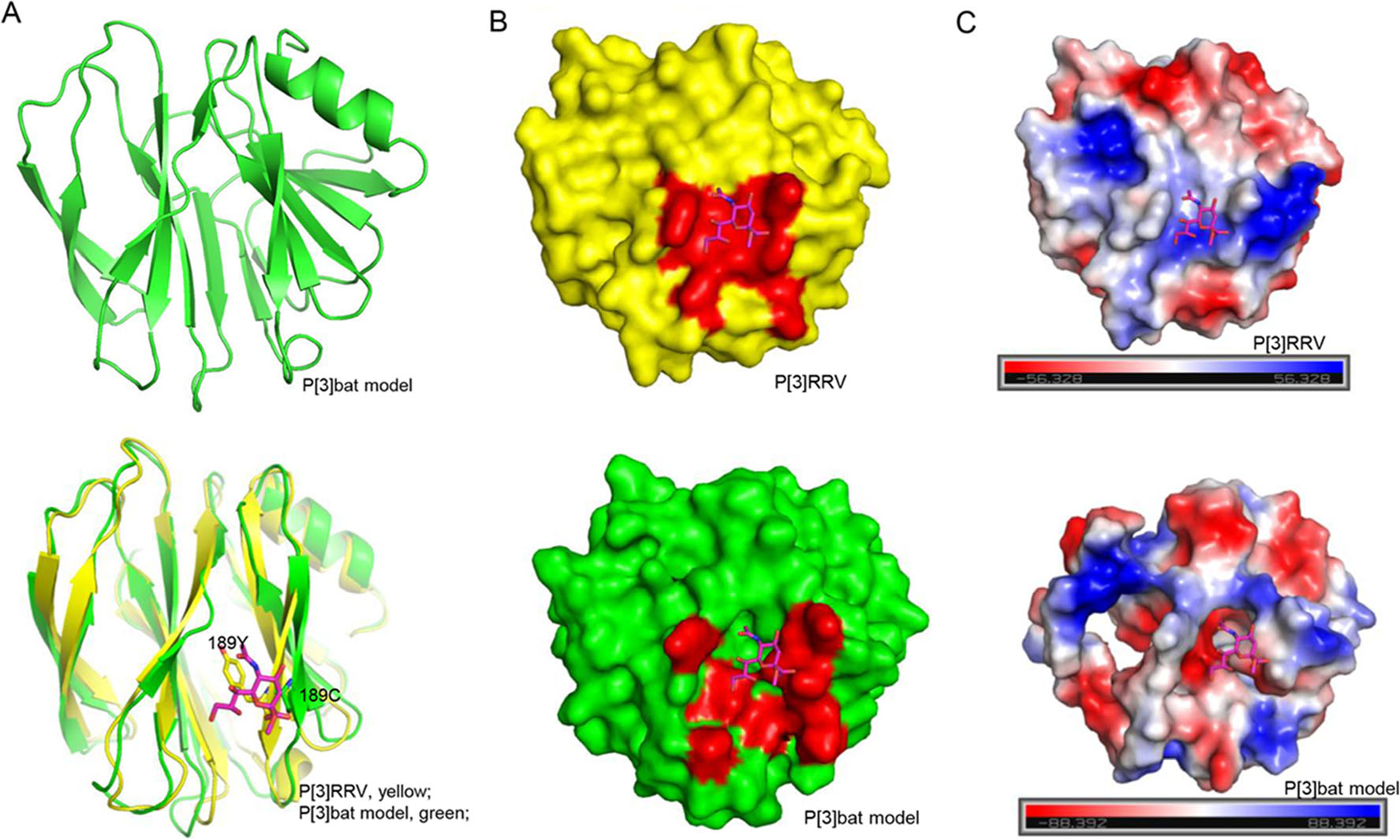
Figure 7. Structural analysis of P[3] VP8*s. A Bat P[3] VP8* model was constructed using the SWISS-MODEL. Bat P[3] VP8* was superimposed to P[3] RRV VP8*s. VP8* was shown as cartoon. Residue 189 Y/C was emphasized using stick. Sialic acid was shown as stick. B P[3] RRV and P[3] bat VP8*s were shown as surface. The glycan binding site and putative glycan binding pocket were colored red. C The electrostatic potential presentation of P[3] RRV and P[3] bat VP8*s. Red and blue indicate the positive and negative charge, respectively.
VP8* Protein Expression and Purification
P[3] VP8*s Recognized Glycans with Different Patterns
Hemagglutination of P[3] VP8*s and Mutants
Sequence Alignment of P[3] VP8*s of Different Origins
Structural Analysis of the Glycan Binding Site of P[3] VP8*s
-
Many rotavirus genotypes can infect different species and species-specific RV strains are frequently identified (Papp et al. 2013). The zoonotic potential has led to the increased burden of RV control and prevention (Doro et al. 2015). The glycan binding specificity of different virus strains are vital for understanding the host infection and tropism (Ramani et al. 2016). VP8*, located at the distal terminal of the VP4 spike, was involved in the glycan recognition and played an important role in the virus attachment (Huang et al. 2012; Ramani et al. 2016). Bats are recognized as the reservoir of many viruses, including the life-threatening Ebola virus, SARS, influenza virus. Rotaviruses in bats were identified by several studies (Esona et al. 2010; He et al. 2013, 2017). P[3] RVs are also found in cats/dogs and some feline/canine-like RVA strains are identified to infect humans sporadically (Tsugawa and Hoshino 2008). The glycan specificity and molecular mechanism of the cross-species transmission is important for understanding the virus prevalence. Here, we delineated the functional features of P[3] RV VP8*s of different species.
Bat LZHP2 P[3] VP8* showed no hemagglutination to all the erythrocytes tested, while other VP8* hemagglutinated the cells, indicating that bat LZHP2 P[3] VP8* probably do not interact with classical ligands that were recognized by Rhesus P[3] RV (Dormitzer et al. 2002a). Glycan binding assay verified that human HCR3A, 12,638, and dog RV198-95 P[3] VP8* bound to glycans with sialic acid while bat P[3] VP8* did not. Human M2-102 hemagglutinated the red blood cells, while showed no obvious binding to the glycans with sialic acid, providing more evidence that human M2-102 was originated from bat P[3] RVs. Human HCR3A and dog RV198-95 P[3] exhibited similar binding to both Neu5Acα2-3Gal and Neu5Acα2-6Gal, indicating that these P[3] RV bound sialic acid irrespective of the α2-3 or α2-6 linkage and possessed the potential of cross-species transmission. These results demonstrated that different P[3] VP8*s had various hemagglutination ability and glycan binding specificity. Bat P[3] VP8*s displayed no binding to sialic acid and probably could not infect human, while human/dog origin P[3] VP8*s bound sialic acid and could infect both human and animals.
Sequence alignment showed that several amino acids were different between bat LZHP2 and human RV VP8*s, including the residue 189. Bat LZHP2 C189Y mutant obtained the ability of hemagglutination while human P[3] Y189C mutant had no hemagglutination, implying that residue 189 was one of the key residues for the ligand recognition and the C189Y alteration in M2-102 RV probably facilitate the sporadic bat-origin RV infection in human. Meanwhile, other amino acids, including residue 144, 155, and 191 may contribute to the different glycan binding capacity of P[3] VP8*.
The structural analysis revealed that the putative glycan binding site in bat P[3] VP8* model was quite different to that in the P[3] RRV VP8*, which partially explained that bat VP8* did not bind sialic acid glycans. These indicated that species barrier probably existed for bat LZHP2 P[3] RVs. Bat LZHP2 P[3] RV could not transmit to humans at present but may infect human sporadically with the C189Y alteration. Analysis of all the reported bat P[3] RV VP8* sequences showed that bat LZHP2 and YSSK5 (He et al. 2017) possessed 189C, while other 8 strains contained 189Y (Fig. 8), implying that some bat P[3] RV strains may have the potential to infect human like the M2-102 strain. Therefore, the consistent surveillance of bat and animal RVs is needed. care and use of laboratory animals were followed.
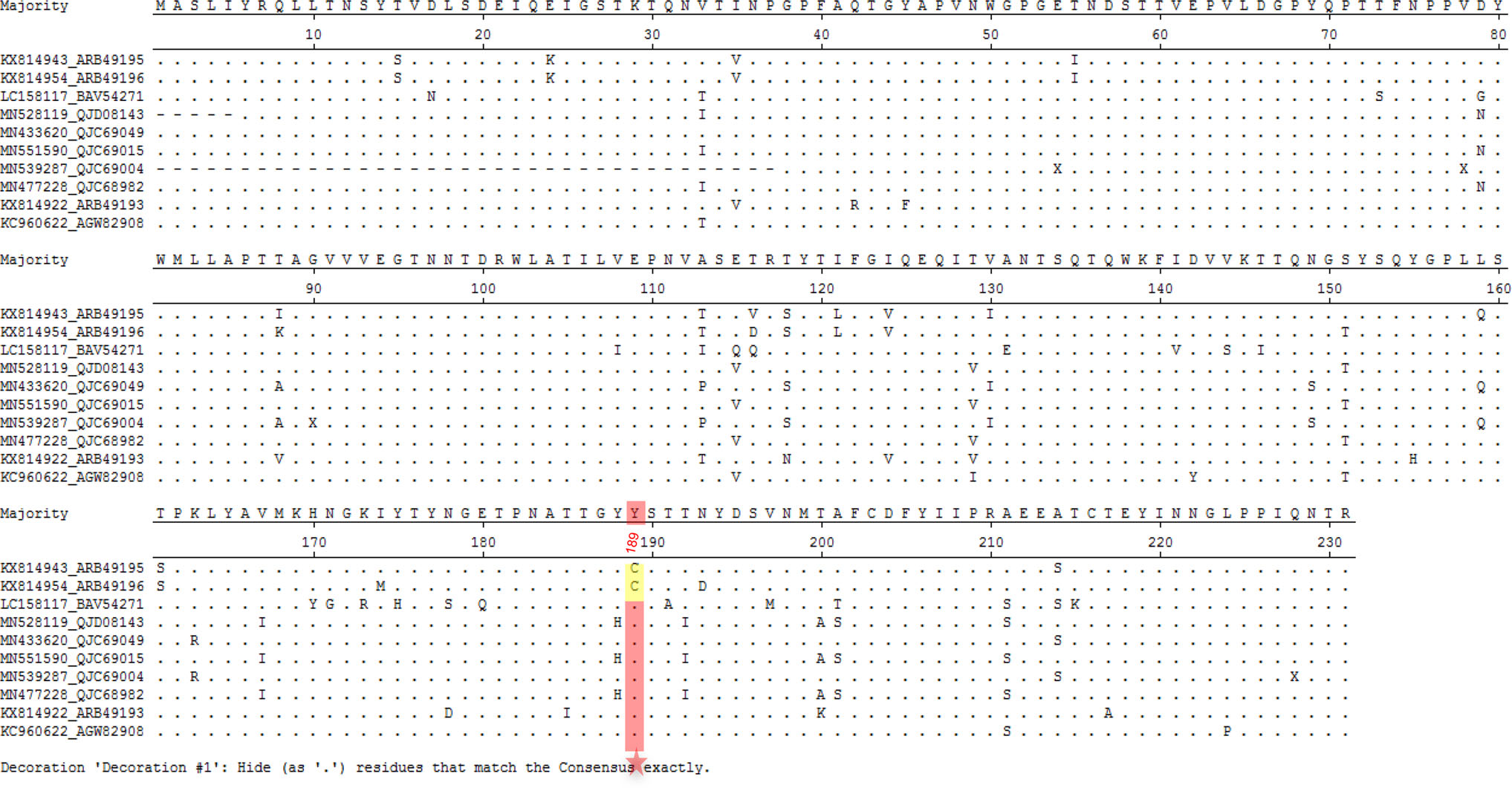
Figure 8. Sequence alignment of ten bat P[3] RV VP8* protein sequences. The sequence alignment was done with MegAlign. Residue 189 was pointed with red star. 189C was colored yellow and 189Y was colored red. GeneBank numbers of the sequences are as following: LZHP2 (KX814943), YSSK5 (KX814954), LUS12-14 (LC158117), GKS-894 (MN528119), BB89-15 (433, 620), GKS-94 (MN551590), BR89-60 (MN539287), GKS-954 (MN477228), BSTM70 (KX814922), MSLH14 (KC960622).
-
We acknowledge the participation of the Protein-Glycan Interaction Resource of the CFG (supporting grant R24 GM098791) and the National Center for Functional Glycomics (NCFG) at Beth Israel Deaconess Medical Center, Harvard Medical School (supporting grant P41 GM103694) for the analysis of samples by glycan microarray. We are grateful to professor Qi Shi and Jia Chen in the National Institute for Viral Disease Control and Prevention for their assistance in the BLI assay. This research was supported by grants from the National Natural Science Foundation of China (NSFC) (No. 21934005) and National Key Research and Development Program of China (2018YFC1200602).
-
ZD, DL, and XS designed the experiments; MXW, DL, TM, and LP performed the protein expression and the glycan binding assay. MXW, MWW, QZ and HW carried out the hemagglutination assay. XS performed the biolayer interferometry assay and the structural analysis. XS and DL analyzed the data and constructed the figures. XS wrote the original draft of the manuscript. DL, ZD and XS revised the manuscript. All authors reviewed the manuscript and provided inputs.
-
All authors declare no potential conflict of interest.
-
All institutional and national guidelines for the care and use of laboratory animals were followed.














 DownLoad:
DownLoad: