HTML
-
Porcine epidemic diarrhea (PED) is a highly contagious enteric disease of swine caused by porcine epidemic diarrhea virus (PEDV). PED is characterized by severe acute watery diarrhea, high morbidity and high mortality, and causing huge economic losses to the pork industry worldwide (Wood 1977; Coussement et al. 1982; Shibata et al. 2000; Lee 2015). PED was first reported in England in 1971, and subsequently in Europe and Asia (Pensaert and de Bouck 1978; Horvath and Mocsari 1981; Pospischil et al. 1981; Takahashi et al. 1983; Kusanagi et al. 1992; Pijpers et al. 1993; Smid et al. 1993; Martelli et al. 2008; Zhou et al. 2012). Later, PED outbreaks in Asia countries were controlled in endemic regions with the widespread use of a prototype CV777-derived vaccine. However, the PEDV variants re-emerged and spread rapidly in China at the end of 2010, and several studies have been reported that the classical strain-derived vaccines failed to protect against the PEDV variants (Li et al. 2012; Sun et al. 2012; Song et al. 2015; Zhang et al. 2020). In April 2013, PEDV variants invaded the United States (US) for the first time. They swept across more than 30 states in the country, causing huge economic losses to the pork industry, and subsequently spread throughout North America, including Canada and Mexico. Afterward, US-like PED epidemics have been reported in South Korea, Taiwan Province of China, and Japan, capturing global attention (Lin et al. 2014; Park et al. 2014; Vlasova et al. 2014; Sasaki et al. 2016). One year later, the recombinant strains generated from the classical and variant PEDV strains were reported in the United States and designated as the S-INDEL strain (Vlasova et al. 2014; Wang et al. 2014). Moreover, the co-infections with the newly emerging S-INDEL strain and the original variant strain have been reported (Vlasova et al. 2014; Su et al. 2018). The S-INDEL strain belongs to G2 genotype together with G2-a and G2-b subgroups, and causes severe diarrhea in newborn piglets, but its pathogenicity is significantly attenuated relative to that of the original virulent PEDV, indicating that PEDV strains are undergoing continuous evolution to adapt to the host environment (Mesquita et al. 2015).
PEDV is an enveloped, single-stranded, plus-sense RNA virus, belonging to the genus Alphacoronavirus Alpha-CoV (CoV). The genome of PEDV is about 28 kb in length, and includes a capped 5′ -untranslated region (5′-UTR), a tailed 3′-UTR, and seven open reading frames (ORF), which encode polymerase peptides pp1a, pp1ab, S protein, accessory protein ORF3, membrane protein (E), membrane protein (M) and nuclear capsid protein (N) (Song and Park 2012). The S glycoprotein on the PEDV surface changed frequently, especially the N-terminal domain of S1 (S1-NTD), and reflects the genetic diversity among the different PEDV strains, allowing researchers to track the genetic dynamics (Lin et al. 2017; Chen et al. 2019). Using reverse genetic techniques, Wang et al. confirmed that the S gene is necessary but not sufficient for the virulence of novel variant PEDV strain BJ2011C (Wang et al. 2018). PEDV with aa deletions in both NTD and the cytoplasmic tail of S protein showed significantly reduced pathogenicity in piglets, confirming that the S protein plays an important role in PEDV virulence (Hou et al. 2017, 2019).
Together with strict biosecurity measures, immunization is vitally important in prevention and control of PED. At present, inactivated vaccines and attenuated live vaccines based on both classical virus strains and virulent epidemic virus strains are available and widely used in China, contributing to the prevention of PED outbreaks. Since the first PED outbreak in the US in 2013, a single inactivated PEDV vaccine and a PEDV subunit vaccine have been conditionally licensed to vaccinate sows. These are thought to provide suckling piglets with passive immunity against PEDV. The above two kinds of vaccines are relatively safe for piglets, but neither provides sufficient passive immune protection (Crawford et al. 2016). Mucosal immunity, especially the secretory IgA (sIgA) component of lactogenic immunity, is deemed to play an important role in the antiviral process. However, attenuated-live vaccines produce higher levels of sIgA than inactivated vaccines, and therefore have tremendous potential utility in the prevention and control of PED (Chattha et al. 2015; Langel et al. 2016). Currently, variant PEDV strains are involved in the continuing epidemic in China. Although the vaccines described above have been widely used in the field, antigenic variations and the insufficient cross-protection afforded by the classical and re-emerged PEDV strains contribute to vaccine failure. Therefore, it is necessary to track the genetic dynamics in PEDV in order to develop the safe and effective vaccines against virulent PEDV strains. In the present study, the virulent FJzz1 strain, previously isolated in our laboratory, was attenuated by serially propagation in vitro. Its biological characteristics and genetic variations were analyzed during its passage, and the pathogenicity of both a low-passage variant and a high-passage variant were investigated to develop potential candidate vaccine strains to effectively prevent PED.
-
Vero E6 cells (African green monkey kidney cells, American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37 ℃ under 5% CO2. PEDV G2 strain FJzz1 (GenBank: MK288006) was previously isolated in Vero E6 cells in our laboratory. A mouse anti-PEDV N protein monoclonal antibody (MAb) was prepared in our laboratory (Wang et al. 2016). An Alexa Fluor 488-conjugated (green) goat anti-mouse secondary antibody was purchased from Life Technologies (Carlsbad, CA, USA), and 4′ 6-diamidino-2-phenylindole (DAPI) was purchased from Beyotime.
-
Vero cells were cultured in a T25 cell culture flask to 90% confluence. The FJzz1 strain stock at the 5th passage (FJzz1-F5) was diluted in 1 mL of serum-free DMEM containing 10 g/mL trypsin at a multiplicity of infection (MOI) of 0.01, and then used to inoculate monolayers of Vero cells that had been rinsed twice with phosphate-buffered saline (PBS). After incubation for 1 h with constant shaking, the virus solution was discarded and the cell culture flask was washed twice with PBS, and 5 mL of maintenance medium containing 10 g/mL trypsin was added. When 80% of the cells had developed a visible cytopathic effect (CPE), the infected cells were lysed with freeze thawing, and centrifuged at 4000 ×g for 1 min at 4 ℃ to harvest FJzz1-F6. The viral stock was then used to inoculate monolayers of Vero cells grown in T25 cell culture flasks. Further propagation was continued in the same way until the 200th passage (FJzz1-F200).
-
Vero cells in six-well cell-culture plates were mock infected or infected with FJzz1 variants F20, F50, F100, F150 or F200 at an MOI of 0.1 as described above. At 24 h postinfection (hpi), the Vero cells were fixed with 80% ice-cold ethanol at 4 ℃ for 1 h. After the cells were washed twice with PBS, mouse anti-PEDV N protein MAb (Wang et al. 2016) and a fluorescein (FITC)-conjugated goat anti-mouse IgG (H + L) antibody were added as the primary and secondary antibodies, respectively. The cell nuclei were stained with DAPI for 15 min at room temperature. The stained cells were washed three times with PBS and analyzed with inverted fluorescence microscopy in the dark.
-
Vero cell monolayers seeded in six-well plates were inoculated with FJzz1 variants F20, F50, F100, F150 or F200 at an MOI of 0.01. After 1 h, the plates were washed three times with PBS, and then covered with prepared liquid agarose solution (2 mL/well) supplemented with 10 μg/mL trypsin at room temperature. The plates were placed in a 37 ℃ cell incubator when the agar had solidified completely. Several days later, when the plaques were clearly visible to the naked eye, the cells were fixed with 4% paraformaldehyde for 2 h at room temperature, and the plaques were visualized by staining with 2.5% crystal violet.
-
Multistep growth curves of FJzz1 variants F20, F50, F100, F150 and F200 were constructed, as previously described, with some modifications. In brief, confluent Vero cells in a 96-well plate were inoculated with FJzz1 variants F20, F50, F100, F150 or F200 at an MOI of 0.01, and the cell culture supernatants were collected every 6 h until 36 hpi. The viral titers of these samples were determined as the 50% tissue culture infectious dose (TCID50), as described below. The confluent monolayers of Vero cells were washed twice with PBS. Ten-fold serial dilutions (10–1 to 10–8) were prepared by adding 100 μL of the test supernatant to 900 μL of DMEM supplemented with 10 μg/mL trypsin, and each dilution was added to confluent monolayers of Vero cells in eight vertical wells of the 96-well plate. The viral CPE was monitored for several days and the viral titers were determined with the Reed and Muench method. Finally the growth curve of each virus was constructed based on the viral titers at different time points after infection.
-
The total RNA of FJzz1 variants F20, F50, F100, F150 and F200 was extracted with the RNeasy Mini Kit (Qiagen, Hilden, Germany), and then reverse transcribed into cDNA with the Revert Aid First Stranded cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA), as described previously. PCR was performed with this cDNA as the template and 16 overlapping pairs of primers (Supplementary Table S1). The genomic sequences of the FJzz1 variants were determined with the next-generation sequencing technology. The aa sequences of the S proteins and the full-length genome sequences of these strains were also aligned using the Clustal W method in MegAlign (DNAStar Lasergene), and the N-linked glycosylation sites in the S proteins of these strains were predicted and analyzed with the NetNGlyc 1.0 Server, as described previously (Chen et al. 2019). A phylogenetic tree was constructed with the neighbor-joining method (NJ) using MEGA 7.0 based on S gene of FJzz1 strain and another 60 reference strains with complete S gene sequences available in GenBank (Supplementary Table S2).
-
Fifteen 5-day-old conventional piglets were confirmed to be negative for PEDV, TGEV, and porcine rotavirus (PoRV) with reverse transcription-PCR (RT-PCR), and negative for PEDV antibodies with IFA using inoculated monolayers of Vero cells. All the animals were raised in the Laboratory Animal Facility at the Shanghai Veterinary Research Institute. The piglets were randomly allocated to three groups, and each group was maintained in a separate room. The piglets in group A or group B were intramuscularly administrated 1 mL of FJzz1-F20 or FJzz1-F200 at a dose of 1.0 × 105 TCID50, respectively. All the piglets in group C were treated with the same amount of DMEM. During the experiment, clinical signs were monitored and recorded daily, including diarrhea, vomiting, anorexia, depression, and changes in body temperature or bodyweight. Fecal consistency was scored every day based on the scale: 0 for solid, 1 for pasty, 2 for semiliquid (mild diarrhea) and 3 for liquid (severe diarrhea), as described previously (Chen et al. 2019). Rectal swabs were collected on days 1, 3, 5, 7, 9, 11 and 13 postinoculation to detect viral shedding. On day 14 postinoculation, all the experimental piglets were euthanized, and the intestinal segments (duodenum, jejunum, ileum, cecum, colon and rectum) were collected for histopathological and immunohistochemical examinations and quantitative analysis.
-
Intestinal segments including jejunum, ileum, cecum and colon were collected and routinely fixed in 10% formalin for 36 h at room temperature. Then the paraffin-embedded tissues were prepared as described previously in some reports (Lin et al. 2015; Liu et al. 2015; Xu et al. 2018). After the paraffin-embedded tissues were cut, deparaffinized, rehydrated, and routinely stained with hematoxylin and eosin (H & E), the sections were scanned for analysis. Sections (5 μm) of formalin-fixed paraffin-embedded tissues were blocked with 1% BSA for 1 h at room temperature, and then incubated with mouse anti-PEDV N protein MAb overnight at 4 ℃, followed by incubation with peroxidase-labeled goat anti-mouse IgG secondary antibody in a humidified chamber for 1 h at room temperature. Finally, the samples were visualized with a 3, 30-diaminobenzidine (DAB) chromogen kit, and counterstain was performed with hematoxylin.
-
The PEDV RNA fecal shedding titers and the viral loads in the intestinal tissues were determined by TaqMan real-time RT-PCR with the probe and primers targeting PEDV N gene as described previously (Kim et al. 2007; Hou et al. 2016; Chen et al. 2019). Relative-quantitative RT-PCR was also performed with TB Green™ Fast qPCR Mix to detect the cytokines such as IFN-α, IFN-λ3, tumor necrosis factor α (TNF-α), interleukin 1β (IL-1β), and IL-8 in the jejunum tissues.
-
All statistical analyses were performed using GraphPad Prism 6 (GraphPad, La Jolla, CA, USA). Statistical significance was assessed with the Student's t-test, and differences were considered significant when the P value was less than 0.05.
Cells, Viruses and Antibodies
Serial Passage of FJzz1 Strain in Vero Cells
Immunofluorescence Assay (IFA)
Plaque Assay
Multi-Step Growth Curve
Genome Sequencing and Genetic Variation Analysis of FJzz1 Variants
Pathogenicity Evaluation of FJzz1 Variants F20 and F200
Histological and Immunohistochemical Staining
RT-qPCR
Statistical Analysis
-
Vero cells were inoculated with FJzz1 variants F20, F50, F100, F150 or F200 at an MOI of 0.01 to analyze the changes in their biological characteristics during the serial passage in vitro. A CPE, characterized by multiple regional cell fusion and syncytium formation, was observed at 10 hpi in the cells inoculated with high-passage variants such as F150 and F200. By contrast, Vero cells inoculated low-passage variants, especially F20, developed visible CPE at 15 hpi, 5 h later than the high-passage variants (Fig. 1A). Vero cells infected with F20, F50, F100, F150, or F200 displayed specific green fluorescence when treated with a Mab directed against PEDV N protein, but no green fluorescence was observed in the uninfected cells (Fig. 1B). All five passages of FJzz1 variants developed visible round plaques on confluent Vero cells in six-well plates. It is noteworthy that the high-passage FJzz1 variants showed higher proliferation than the low-passage variants, which contributed to larger size form of plaques (0.21–0.26 mm) in Vero cells infected with the high-passage FJzz1 variants than those with the low-passage variants (0.08–0.12 mm) (Fig. 1C). The multistep growth kinetics of F20, F50, F100, F150, and F200 based on the TCID50 at different hpi were visualized, and showed that the viral titers of the low-passage variants (F20, F50, and F100) peaked at 30 hpi, whereas those of the high-passage variants (F150 and F200) peaked at 24 hpi, 6 h before the low-passage variants (Fig. 1D). In conclusion, these results indicate that the sensitivity and adaptability of the FJzz1 variants to Vero cells gradually increased during their serial passage in vitro.
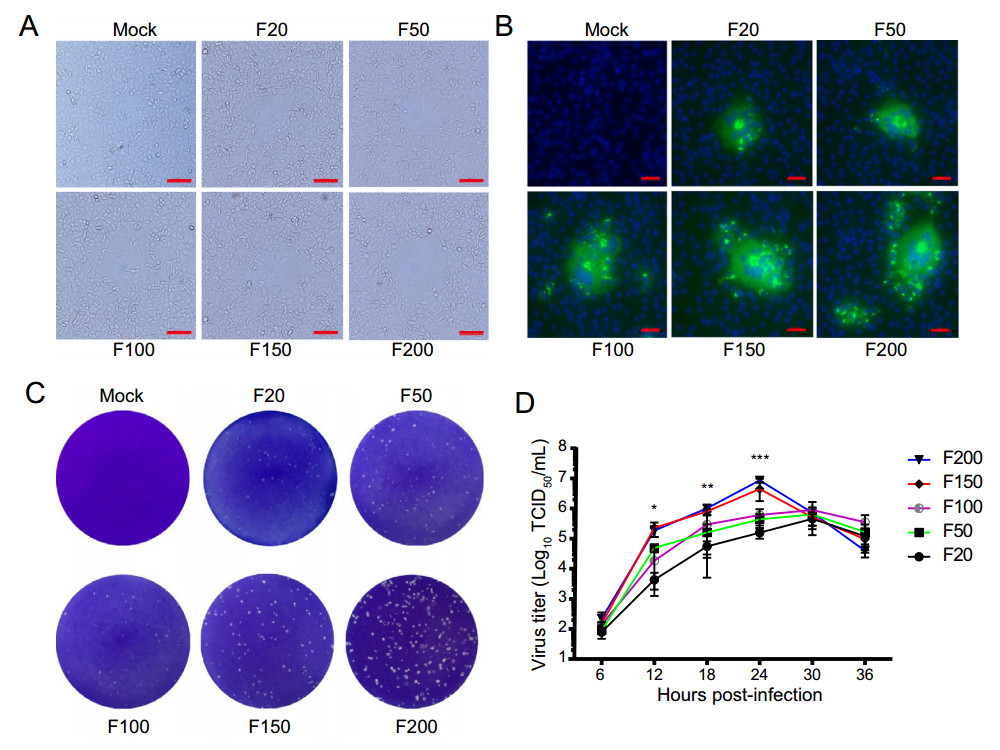
Figure 1. Biological characterization of FJzz1 variants during serial passage in vitro. A Vero cells were mock infected or infected with FJzz1 variants F20, F50, F100, F150, or F200 at an MOI of 0.01, and a CPE was observed at 24 hpi. Scale bar = 100 µm. B Monolayers of Vero cells inoculated with FJzz1 variants F20, F50, F100, F150, or F200 were tested with an IFA using a MAb to PEDV N protein. Scale bar = 50 µm. C Crystal-violet-stained plaques formed on the monolayers of Vero cells inoculated with FJzz1 variants at different passages at 3 dpi. D Vero cells were infected with FJzz1 variants at different passages at an MOI of 0.01. The cell lysates were collected at the designated times and titrated with a TCID50 infectivity assay. Asterisk (*) indicates a significant difference between FJzz1-F20 and FJzz1-F200 (*P < 0.05; **P < 0.01; ***P < 0.001).
-
To further analyze the genetic variation in the FJzz1 variants, the full-length genomic sequences of these variants were aligned using Clustal W in MegAlign, and the results showed that relative to F20, the other passaged variants (F50, F100, F150, and F200) had 7, 14, 18 and 19 aa changes, respectively (Table 1), indicating that genomic sequences of the FJzz1 variants tended to be stable during serial passage. Different degrees of aa changes were detected in the ORFs of PEDV, including ORF1a, ORF1b, S, ORF3 and M. F100, F150 and F200 showed the same numbers of aa changes in Nsp2, ORF3 and M, which accounted for 0.13%, 0.44% and 0.44% of their total aa, respectively. The change rate of the S protein in FJzz1 variants increased during their serial propagations in vitro, reaching the peak change rate (1.01%) in F200. Notably, the aa changes in FJzz1 variants were mainly concentrated in the S protein, with high change rates ranging from 72.22% to 85.71% of the total changed aa (Fig. 2A), whereas other proteins (Nsp2, Nsp3, Nsp13, ORF and M) had lower aa change rates. The changed aa of the S protein in the FJzz1 variants were scattered in both the S1 and S2 subunits, which is consistent with the differences observed between the classical strain and its attenuated variants and between other mutant epidemic strains and their attenuated variants (Fig. 2B). In addition, a phylogenetic tree was constructed based on the S gene of the FJzz1 strain and another 60 reference strains for which the complete S gene sequences were available in GenBank. The phylogenetic analysis showed that the FJzz1 strain and the 60 reference strains formed two genogroups: the G1 genotype mainly contained the classical strains, such as CV777 and DR13, whereas the G2 genotype contained the FJzz1 strain and other variant pandemic strains (Fig. 2C). All the aa changes in the FJzz1 variants at different passages are listed in Table 2.
ORFs Encoded proteins F20 F50 F100 F150 F200 ORF1a Nsp2 0 (0)* 0 (0) 1 (0.13) 1 (0.13) 1 (0.13) Nsp3 0 (0) 1 (0.06) 1 (0.06) 1 (0.06) 1 (0.06) ORF1b Nsp13 0 (0) 0 (0) 0 (0) 1 (0.17) 1 (0.17) S (1386) S 0 (0) 6 (0.43) 12 (0.87) 13 (0.94) 14 (1.01) ORF3 (225) ORF3 0 (0) 0 (0) 1 (0.44) 1 (0.44) 1 (0.44) M (227) M 0 (0) 0 (0) 1 (0.44) 1 (0.44) 1 (0.44) Total number 0 7 14 18 19 *Number of aa changes and change rates of the corresponding proteins (%) are indicated in parentheses. Table 1. Statistics of aa change numbers and change rates in the corresponding proteins at different passages.

Figure 2. Phylogenetic analysis of FJzz1 strain. A Rate of mutation was calculated as the percentage of changed aa in the corresponding protein, including Nsp2, Nsp3, Nsp13, ORF3, S, and M. B Positions of the changed aa in the S protein are shown. The y-axis represents the positions of the changed aa, and the x-axis represents the six different pairs of virulent parental /attenuated strains (Pair 1: DR13/attenuated DR13; Pair 2: PC22A/PC22A-P160; Pair 3: 83P-5/83P-5-100th; Pair 4: KNU-141112-P5/KNU-141112-S-DEL2-ORF3; Pair 5: YN15/Y144; Pair 6: FJzz1-F20/FJzz1-F200). C A phylogenetic tree was constructed based on S gene of FJzz1 strain and another 60 reference strains with complete S gene sequences available in GenBank. The PEDV strain FJzz1 was marked with the filled triangle.
ORFs Amino acid position F20 F50 F100 F150 F200 ORF1a Nsp2 (750) 699 M M L L L Nsp3 (1656) 1562 M I I I I ORF1b Nsp13 (597) 968 A A A T T S (1386) 42 Q Q H Q Q 55–57 IGE K– K– K– K– 128 F F Y Y Y 265 D D D A A 378 D D N N N 490 T T R R R 773 K K K K N 877–878 SG RR RR RR RR 900 L L L L V 1009 N N D D D 1338 I I T T T 1353 C F F F F ORF3 (225) 170 Y Y H H H M (227) 159 G G D D D Table 2. Amino acid changes that occurred at different passages.
-
The S glycoprotein is one of the essential structural proteins of CoVs, and plays a vital role in genetic variation, viral invasion, viral virulence, and the production of neutralizing antibodies. To explore the antigenic variation in the FJzz1 variants during their serial passage, a sequence alignment of the deduced S glycoprotein aa in different generations was constructed. This showed that compared with the classic strain CV777, the low-passaged FJzz1 variants (F5 and F20) contained five aa insertions. However, after serial passage in vitro, the high-passaged FJzz1 variants (F50, F100, F150 and F200) showed characteristic consecutive aa deletions in the S1-NTD region (55I56G57E → 55K56△57△), and all the FJzz1 variants in different generations had two aa deletions relative to CV777 (Fig. 3). Interestingly, an aa change (N-D) occurred in variants F100, F150 and F200, but not at the same position in variants F5, F20, and F50, resulting in the disappearance of a predicted N-linked glycosylation site in the S glycoprotein in variants F100, F150 and F200. A total of six consistent aa changes were observed at the epitopes in COE and SS6, but the effects of these changes on the neutralization activity of these epitopes remain to be investigated. CoVs contain two conserved motifs, KVHVQ and YxxΦ, in the cytoplasmic tail of the S protein, which may affect the pathogenicity of CoVs (Schwegmann-Wessels et al. 2004; Youn et al. 2005; Winter et al. 2008). In the present study, we noted that these two motifs were conserved and stable during the serial passage of FJzz1.
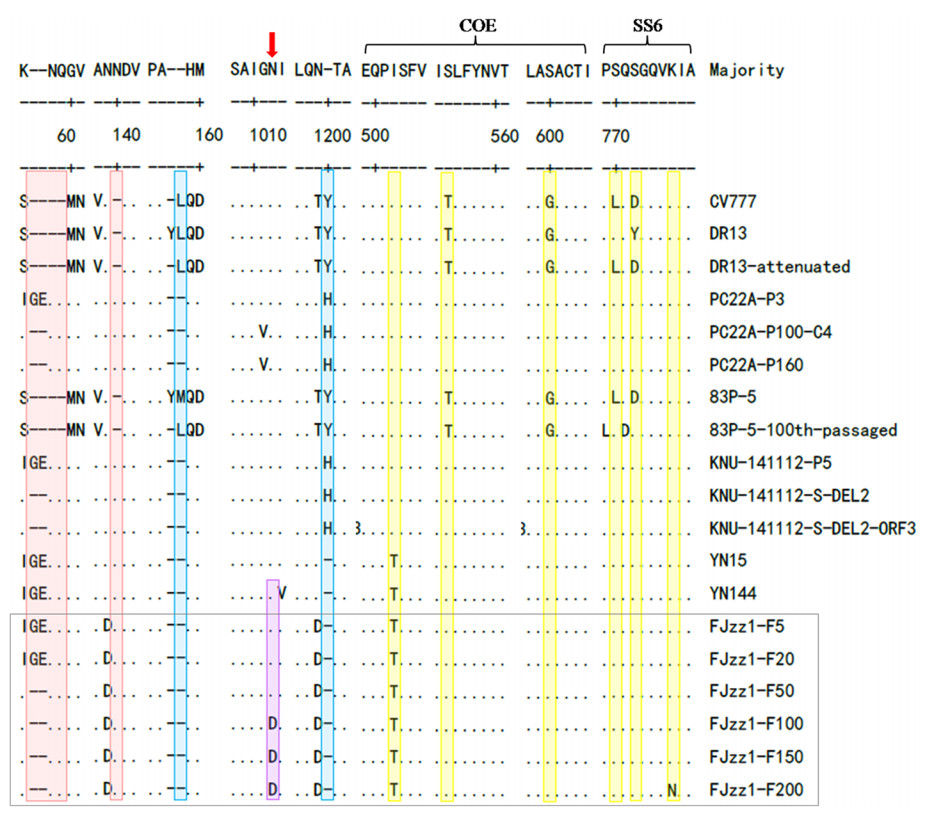
Figure 3. Alignment of S protein sequences of FJzz1 variants at different passages and other PEDV strains. S protein aa sequences of FJzz1 variants at different passages (marked with black box) and other PEDV strains including the classical CV777 strain, were aligned using Clustal W. Predicted N-linked glycosylation sites in the FJzz1 variants are marked with red arrows. Regions inserted and deleted relative to CV777 are highlighted in red and blue, respectively. The substitutions in neutralizing epitopes are highlighted in yellow.
-
To determine whether the virulence of the FJzz1 strain changed during its serial passage in vitro, FJzz1-F20 and FJzz1-F200 were analyzed simultaneously to evaluate their pathogenicity in suckling piglets. The results showed that piglet A2 in the group infected with FJzz1-F20 (group A) began to show diarrhea at 15 hpi, with typical clinical signs such as loose yellowish stools and loss of appetite. Most of piglets in group A developed diarrhea by 24 hpi, and all five piglets inoculated with FJzz1-F20 developed severe watery diarrhea within 48 h. However, the clinical signs of piglet A3 were alleviated after 8dpi, with an increase in appetite. By contrast, most of the piglets infected with the same dose of FJzz1-F200 were in good condition, and their activity levels and feed intake were significantly higher than those of the piglets in group A. Piglets B4 and B5 in the group infected with FJzz1-F200 (group B) showed transient mild diarrhea signs at 2 and 6 days postinfection (dpi), respectively, lasting for less than 24 h. The other piglets in group B showed no obvious signs, and none of the piglets in the control group (group C) showed any clinical signs throughout the experiment, consistent with expectations (Fig. 4A). The daily body temperature and bodyweight of each piglet were monitored, and the body temperatures of most piglets remained relatively stable, except for the moribund piglets, whose body temperatures dropped significantly (Fig. 4B). There were no significant differences in the bodyweights of the piglets among the groups before inoculation. However, the bodyweights of the piglets in group A decreased significantly within 6 dpi (Fig. 4C). By contrast, the bodyweights of most piglets in group B remained relatively stable, and increased significantly after 1 week. The weights of all the piglets in group C maintained a gradual increase throughout the experiment, as expected. Piglets A2, A5, A4, and A1 in group A gradually became moribund and died or were euthanized at 2, 4, 5, and 7 dpi, respectively. The mortality rate of group A was 80%, whereas no mortality was observed in the piglets of group B or group C (Fig. 4D). These results indicate that FJzz1-F20 was virulent in piglets, whereas FJzz1-F200 was significantly attenuated after serial passage in vitro.
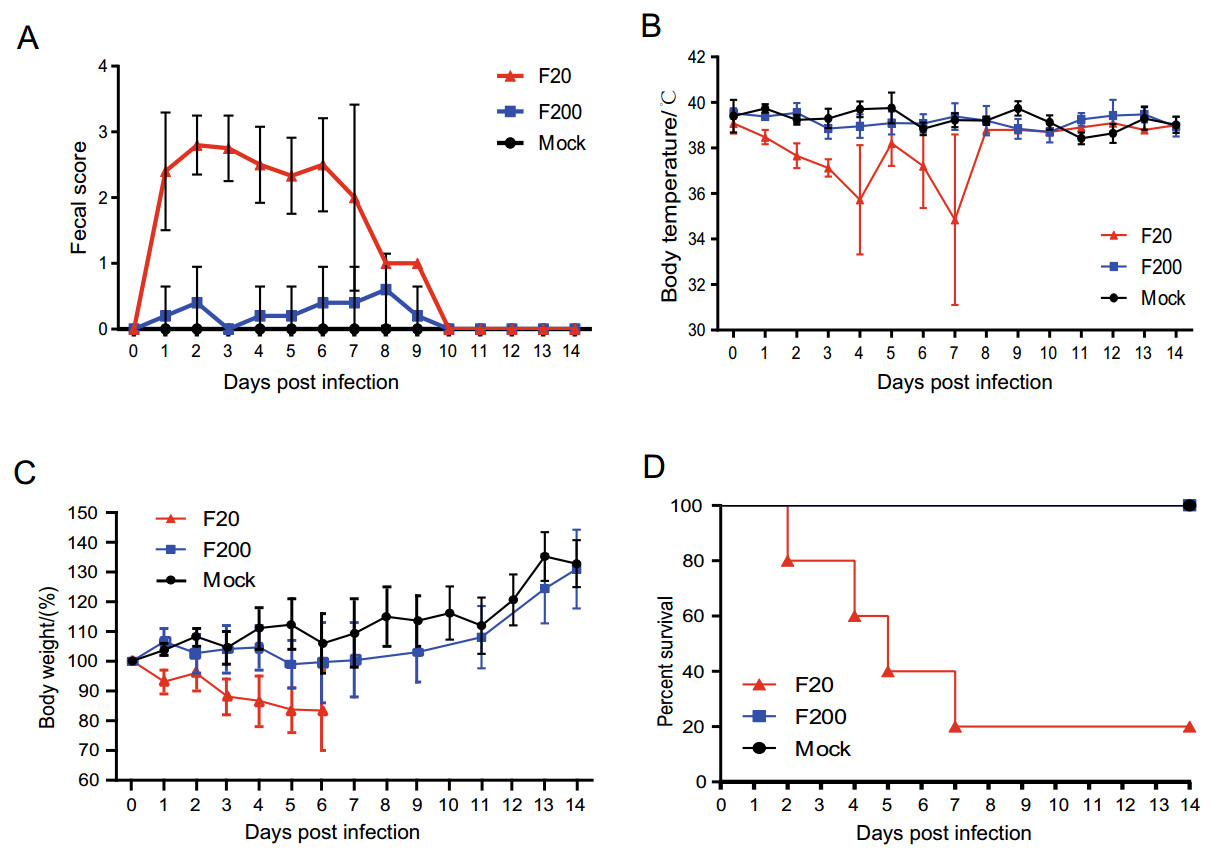
Figure 4. Pathogenicity analysis of FJzz1-F20 and FJzz1-F200. A Fecal scores of piglets with the valuation standard: 0 = normal; 1 = soft; 2 = semi-fluid; 3 = watery diarrhea. B Changes in average body temperature in each group within the first 14 dpi. C Average bodyweight changes in each group. D Survival rates of piglets in each group. Survival curves of piglets infected with DMEM (Mock), FJzz1-F20, or FJzz1-F200 are shown.
-
Pathological and histological examinations were performed to systematically and intuitively evaluate the pathogenicity of variants FJzz1-F20 and FJzz1-F200 in suckling piglets. FJzz1-F20 infection caused severe pathological changes throughout the entire small intestine characterized by thin or even transparent intestinal walls and a large amount of yellowish fluid in the intestinal cavity. Varying degrees of swelling and bleeding were observed in the inguinal lymph nodes and mesenteric lymph nodes, which appeared dark red or dark purple, whereas other organs such as the lung, spleen, and kidney, showed no visible pathological changes (Supplementary Figure S1). By contrast, the piglets in the FJzz1-F200 infection group and the control group showed no visible pathological lesions in these organs, as expected. Histological staining revealed that FJzz1-F20 infection caused severe histopathological lesions in all the intestinal segments, especially the jejunum and ileum, characterized by the atrophy, shortening, or even shedding of the intestinal villi (Fig. 5A). The large intestine, particularly the cecum, was slightly damaged, showing varying degrees of atrophy. In the FJzz1-F200 infection group, no visible microscopic lesions were detected other than slight intestinal villus damage in the jejunum. The intestinal villi of the piglets in the control group were intact with no microscopic lesions, as expected. Staining for the PEDV N protein appeared as tiny stipples within the cytoplasm of the jejunal and ileal cells. PEDV-positive enterocytes were sporadically detected in the jejunum and ileum in both the FJzz1-F20- and FJzz1-F200-infected piglets, but the amount of PEDV detected was significantly higher in the FJzz1-F20-infected group than in the FJzz1-F200-infected group, whereas no PEDV was detected in the control group (Fig. 5B). In conclusion, these results show that the pathogenicity of FJzz1-F200 was significantly lower than that of FJzz1-F20.
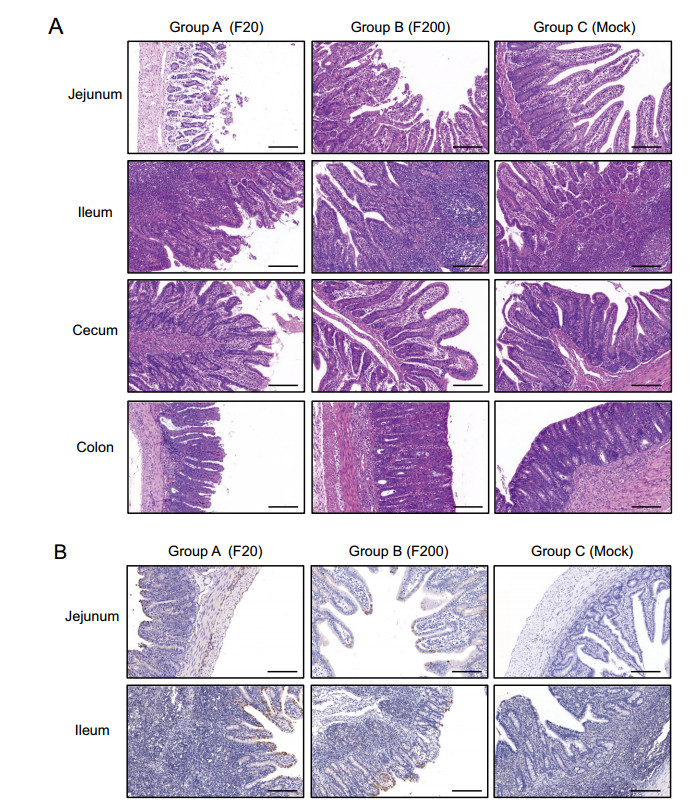
Figure 5. Histopathological lesions in different intestinal segments of piglets inoculated with FJzz1-F20 or FJzz1-F200. A Different intestinal segments (Jejunum, Ileum, Cecum, and Colon) from FJzz1-F20-, FJzz1-F200- or mock-inoculated piglets were collected on the day of death or at the final time points for hematoxylin–eosin staining. B Immunohistochemical detection. Jejunal and Ileal tissues from each group were stained with a monoclonal antibody directed against PEDV N protein (1:200 dilution), scale bar = 200 μm.
-
Viral shedding in the feces was measured with TaqMan real-time RT-PCR (Kim et al. 2007). The viral shedding titers peaked within 1–3 dpi, in a range of 3.24 × 108 to 1.13 × 109 copies/mL, after which they decreased, ranging from 2.32 × 104 to 6.07 × 106 copies/mL at 5–13 dpi (Fig. 6A). However, the piglets in group B shed virus at a low level in the initial stage of infection, after which the titers increased gradually to reach a peak of 7.83 × 106 copies/mL at 7 dpi, significantly lower than that in the piglets of group A. The Viral shedding titers in the piglets in group B then decreased again until it did not differ significantly from that of group A (Fig. 6A). We simultaneously examined the viral loads in the different segments of the intestines, including the ileum, jejunum, cecum, and colon. As shown in Fig. 6B, all the segments of the intestine showed high viral loads in the piglets of group A, ranging from 1.11 × 106 to 6.56 × 108 copies/g, significantly higher than those in the piglets of group B (8.26 × 104 to 1.36 × 105 copies/g). During the whole experiment, both the fecal viral shedding rate and the viral loads in the different segments of intestines were deemed to be negative in group C (Fig. 6B). These results confirm that the pathogenicity of FJzz1-F200 was significantly attenuated in piglets relative to that of FJzz1-F20.

Figure 6. Fecal viral shedding and quantification of viral load in different intestinal segments. Viral shedding in feces A and the viral loads in different intestinal segments B were determined with TaqMan real-time RT-PCR targeting the PEDV N gene. Asterisk (*) indicates a significant difference between FJzz1-F20 and FJzz1-F200 (*P < 0.05; **P < 0.01; ***P < 0.001).
-
The production of cytokines including types I/III IFN and pro-inflammatory cytokines in the target tissue is part of the innate immune response to viral infection. To investigate the innate immune responses induced by PEDV infection in vivo, the cytokines in the jejunums of the piglets were quantified with relative RT-qPCR. The levels of the type I IFN (IFN-α) and type III IFN (IFN-λ3) transcipts induced in the jejunum tissue were significantly higher after FJzz1-F20 and FJzz1-F200 infection than in the control group, and FJzz1-F200 infection induced higher levels of IFN than FJzz1-F20, especially IFN-λ3, which was significantly elevated (Fig. 7). FJzz1-F20 infection also induced higher IL-1β and IL-8 in the transcript levels in the jejunum tissue than were observed in the control group. FJzz1-F20 infection induced higher levels of IL-8 mRNA than did FJzz1-F200 infection. Moreover, compared with the control group, FJzz1-F200 infection significantly increased the transcription levels of pro-inflammatory cytokines, such as TNF-α, IL-1β, and IL-8, in the jejunum tissue. These results suggest that PEDV infection induces the innate immune response in its target tissues, inhibiting viral replication.

Figure 7. Quantification of cytokines in jejunal tissue. Production of cytokines, including A type I interferon (IFN-α) and B type III interferon (IFN-λ3), and pro-inflammatory cytokines, such as C TNF-α, D IL-1β and E IL-8, were measured with real-time RT-PCR in jejunal tissue. The experiment was performed three times and the representative data are shown. Error bars represent means ± standard deviations (SD). *P < 0.05; **P < 0.01; ***P < 0.001.
Biological Characteristics of FJzz1 Variants During Serial Passage
Genetic Variation Analysis of FJzz1 Variants
Structural Analysis of S Glycoproteins of FJzz1 Variants
Clinical Signs in Suckling Piglets Infected with FJzz1 Variants
Histopathological Lesions in Suckling Piglets Infected with FJzz1 Variants
Viral Shedding in Feces and Viral Loads in Different Intestinal Segments
Cytokines Induced in the Jejunum by FJzz1 Infection
-
In recent years, outbreaks of PED caused by highly virulent PEDV variants have frequently re-emerged in China, causing devastating economic losses to the pork industry (Li et al. 2012; Sun et al. 2012). PEDV is a member of the genus Alphacoronavirus, which includes enveloped, single-stranded, positive-sense RNA viruses. To understand the genetic dynamics and the changes in its pathogenicity during its passage in vitro, FJzz1-F5, an epidemic PEDV variant strain that was isolated and purified previously by our laboratory, was serially propagated in Vero cells for up to 200 passages. The biological characteristics and genetic variations in the FJzz1 variants at different passages, including F20, F50, F100, F150, and F200, showed that PEDV's susceptibility and adaptability to Vero cells increased gradually as it was serially passaged, and that the genomic sequence tended to be stable. Different levels of aa changes were detected in the ORFs of the FJzz1 variants, including ORF1a, ORF1b, S, ORF3 and M. Among these, the Nsp2, ORF3 and M aa sequences tended to be more stable in the high-passage FJzz1 variants than in the low-passage FJzz1 variants. The aa changes of FJzz1 variants F50, F100, F150, and F200 relative to F20, were mainly concentrated in the S glycoproteins. A phylogenetic tree based on the full-length S gene indicated that the FJzz1 strain belonged to the G2-b subgroup. Notably, F5 and F20 displayed five aa insertions in the S1-NTD region relative to the same region in CV777. However, with serial passage in vitro, two reversed aa changes were detected in the high-passage FJzz1 variants (F50, F100, F150, and F200). This phenomenon has also been observed in other variant epidemic strains and their attenuated counterparts (Sato et al. 2011; Park et al. 2012; Chen et al. 2015; Lee et al. 2017; Lin et al. 2017). The N-linked glycosylation site in the S glycoprotein of severe acute respiratory syndrome CoV (SARS-CoV) has previously been shown to be essential for viral entry (Han et al. 2007). Another study reported that a variant PEDV had stronger sugar-binding activity than that of classical CV777 (Deng et al. 2016). During the serial passage of the FJzz1 strain, an aa mutation (N-D) occurred in variants F100, F150, and F200, but not at the same position in variants F5, F20, or F50, causing the disappearance of a predicted N-linked glycosylation site in the S glycoprotein of variants F100, F150, and F200. We speculate that this disappearance of the predicted N-glycosylation site, as well as other changes of predicted N-glycosylation sites in variant epidemic strains relative to the classical CV777 (previously reported by our laboratory) may affect the recognition between the specific receptor, a sugar coreceptor, or other unknown coreceptor of the variant PEDV and the receptor-binding domains in S that are considered to influence the invasion efficiency of PEDV. Mutations of the predicted N-glycosylation sites may affect the antigenicity of some vital epitopes, which can explain, at least to some extent, why CV777-derived vaccines do not provide complete cross-protection against the re-emerged virulent PEDV strains. Neutralizing epitopes also induce specific neutralizing antibodies in host cells, playing an important role in the prevention and control of PED. In the present study, the FJzz1 variants at different passages showed six consistent aa changes relative to CV777, at epitopes in COE and SS6. Whether these changes affect the neutralization activity of the epitopes remains to be investigated. However, these adaptive changes may also be an evolutionary strategy of PEDV to escape the immune responses caused by vaccination, which should be confirmed by further experiments, including reverse genetic analyses.
Our laboratory previously evaluated the pathogenicity of FJzz1-F5 in piglets after its administration via oral and intramuscular routes. We found that both infection routes resulted in typical clinical signs in suckling piglets, confirming that the variant PEDV isolate FJzz1-F5 is highly pathogenic in suckling piglets (Chen et al. 2019). The S gene of CoVs is an important target gene for investigating genomic function, because it tends to vary with genetic diversity (Sun et al. 2012; Wu et al. 2012; Wang et al. 2014). The development of the reverse genetic manipulation technology allows us to explore the pathogenesis of PEDV and to rationally design live-attenuated vaccines against it (Hou and Wang 2019). In a previous study, a rescued virus with a 197-amino-acid deletion in NTD-S1 (icPC22A-S1Δ197) was attenuated relative to the highly virulent PEDV strain PC22A in neonatal piglets. However, icPC22A-S1Δ197 did not provide complete cross-protection against challenge with PC21A, suggesting that this 197-amino-acid region contains important epitopes for inducing protective immunity (Hou et al. 2017). Another study confirmed that the S gene is necessary but not sufficient for the virulence of PEDV, and that the structural-protein-coding region and 3′-UTR contribute to the virulence of PEDV (Wang et al. 2018). In the present study, the clinical signs and histopathological lesions in the suckling piglets indicated that the low-passage FJzz1-F20 was highly pathogenic to suckling piglets. By contrast, most of piglets in the FJzz1-F200-infected group had no obvious clinical signs throughout the whole experiment, except for mild transient (< 24 h) diarrhea in two piglets, suggesting that the high-passage FJzz1-F200 was significantly attenuated. Moreover, the viral shedding titers in piglets infected with FJzz1-F20 peaked at 24 hpi, 6 days earlier than in piglets infected with FJzz1-F200, and the peak shedding titers was 143 times higher in the group A piglets than in the group B piglets. Furthermore, the intestinal tissues of the piglets infected with FJzz1-F20 has viral loads 4820 times higher than those of the piglets infected with FJzz1-F200, confirming that the pathogenicity of FJzz1-F200 in piglets was significantly attenuated relative to that of FJzz1-F20 after its serial passage in vitro. PEDV entry is mainly mediated by S glycoprotein, a key structural protein that is responsible for both the host range and the tissue tropism of the virus. Researchers have demonstrated that S gene is susceptible to mutation, thus varying the pathogenicity of PEDV (Hou et al. 2017; Suzuki et al. 2018; Wang et al. 2018). In the present study, a total of 19 changed aa were observed in the ORFs of PEDV, including ORF1a, ORF1b, S, ORF3, and M, and the S protein displayed the highest change rate, with 14 changed aa, which accounted for 73.68% of the total changes in PEDV. We speculated that these changed aa may be associated with the difference in pathogenicity of low-passage FJzz1-F20 and high-passage FJzz1-F20, and further experiments, including reverse genetics analyses, are required to confirm this. Many CoVs contain the conserved motifs KVHVQ and YxxΦ in the cytoplasmic tail of the S protein, and their functions varied in different CoVs (Youn et al. 2005; Ujike et al. 2016). Hou et al. demonstrated that the Vero-cell-adapted PEDV variants lacking functional KVHVQ and YxxΦ motifs were attenuated in piglets, suggesting that these motifs are key viral virulence factors (Hou et al. 2019). However, in the present study, we found no any changes in these two conserved motifs, indicating that the KVHVQ and YxxΦ motifs are necessary but not sufficient for the virulence of PEDV, which must be a multigenic event.
Pro-inflammatory cytokines and IFNs, especially type I/III IFNs represent the first line of the host immune defenses, playing a pivotal role in the antiviral response. Researchers have demonstrated that both the N and E proteins of PEDV trigger endoplasmic reticulum stress and activate NF-κB, which is responsible for the upregulation of IL-8 expression (Luppi et al. 2007; Xu et al. 2013; Ding et al. 2014). In this study, we found that FJzz1-F20 infection induced higher IL-1β and IL-8 transcription in jejunum tissues than was observed in the control, and higher levels of IL-8 transcription than after FJzz1-F200 infection. Similarly, FJzz1-F200 infection significantly increased the transcription levels of TNF-α, IL-1β, and IL-8 in jejunal tissues relative to those in the control group. These pro-inflammatory cytokines exert their antiviral effects by regulating the immune response and the inflammatory reaction of host, either directly or indirectly. Recent studies have demonstrated that type III IFN induces higher levels of interferon-stimulated genes (ISG) to inhibit PEDV replication than type I IFN (Li et al. 2017, 2019; Zhang et al. 2018; Deng et al. 2019). The results of the present study show that both FJzz1-F20 and FJzz1-F200 infection induced higher levels of type I IFN (IFN-α) and type III IFN (IFN-λ3) than were observed in the control group, and FJzz1-F200 infection induced high levels of IFN transcription than FJzz1-F20 infection, especially that of IFN-λ3, which differed significantly in the two groups. Therefore, we conjecture that the high level of IFN-λ3 induced by FJzz1-F200 infection might inhibit PEDV proliferation in the small intestinal tissues, which is consistent with the attenuated pathogenicity of FJzz1-F200 in piglets that displayed clinical signs of PEDV infection. These results suggest that the host activates a series of immune responses to resist viral invasion, but the specific mechanism is not fully understood. Therefore, it will be necessary to analyze the molecular basis of the genetic variation and pathogenicity of variant epidemic PEDV strains from the perspective of the host.
In conclusion, our study demonstrates that fecal viral shedding and the viral load in different segments of the intestines were significantly lower after infection with high-passage FJzz1-F200 than after infection with low-passage FJzz1-F20. Moreover, no obvious histopathological lesions were observed in the FJzz1-F200-infected group, although PEDV-positive enterocytes were sporadically detected within the cytoplasm of jejunal and ileal cells, indicating that FJzz1-F200 replicated slowly in the intestinal tissues of the infected piglets. FJzz1-F200 also induced high leveld of innate-immune-response components, including type I and type III IFNs and pro-inflammatory cytokines, in the target tissues of the infected piglets. We speculated that the attenuated FJzz1-F200 may have the potential to be a candidate for developing PEDV live attenuated vaccines, and such more experiments as challenge study need to be done to evaluate the protection in the future.
-
The study was supported by the National Program on Key Research Project of China (2016YFD0500100), the Shanghai Youth Scientific and Technological Yang Fan Program Grant (20YF1457800), the National Natural Science Foundation of China (31472207), the earmarked fund for Modern Agro-industry Technology Research System of China (CARS-36), the China Postdoctoral Science Foundation (2020M670555) and Shanghai Minhang District talent development special funds.
-
YJZ and GZT conceived and designed the experiments. PFC, XWZ, STZ and TXZ performed the experiments. PFC, XMT and XW analyzed the data. WT, FG, and LXY contributed reagents/materials/analysis tools. YFJ, HY, and ZBY participated in part of experimental work. PFC wrote the paper. YJZ checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
All animal experiments were approved by the Ethical Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Shvrip-20180320).
















 DownLoad:
DownLoad: