HTML
-
The Baculoviridae are a large family of insect-specific viruses with circular, covalently closed, double-stranded DNA genomes of 80–180 kb in size and encoding of 89–183 genes (Rohrmann 2019). Based on their genome sequences, baculoviruses can be divided into four genera: Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus (Jehle et al. 2006). Alphabaculoviruses can be further subdivided into Group I and Group II viruses (Herniou et al. 2001). The most notable differences between these two groups are that Group I nucleopolyhedroviruses (NPVs) use GP64 as their budded virus (BV) fusion protein, whereas Group II NPVs lack GP64 and use the F protein (Pearson and Rohrmann 2002). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the archetype species of Alphabaculovirus.
Baculovirus infection produces two distinct viral phenotypes: BVs and occlusion-derived viruses (ODVs) (Rohrmann 2019). BVs are responsible for spreading infection within susceptible insect cells and tissues, whereas ODVs initiate primary infection in the midgut epithelia of infected insects and are transmitted among insects (Slack and Arif 2006). Transcription and replication of viral DNA and assembly of nucleocapsids occur in a structure called the virogenic stroma (VS) (Fraser 1986; Young et al. 1993). Synthesized nucleocapsids are transported from the VS to the ring zone and then egress from the nucleus to the cytoplasm, and bud from the plasma membrane to form BVs. Subsequently, nucleocapsids retained in the ring zone of the nucleus are enveloped by intranuclear microvesicles to form ODVs, which are then embedded within the polyhedrin to form OBs (Rohrmann 2019).
Most DNA viruses, including herpesviruses and baculoviruses, replicate and assemble their nucleocapsids in the nucleus (Johnson and Baines 2011; Rohrmann 2019). Egress of nucleocapsids is indispensable for the formation of mature virions and viral pathogenicity. This process also represents a good target for disrupting viral infection. The mechanism through which herpesvirus nucleocapsids egress has been well characterized (Johnson and Baines 2011; Hellberg et al. 2016). By contrast, the mechanism of baculovirus nucleocapsid egress remains unclear. According to previous reports, host proteins including the actin cytoskeleton, N-ethylmaleimide-sensitive fusions proteins and endosomal sorting complex are required for transportIII (ESCRT-III) (Guo et al. 2017a; b; Ohkawa and Welch 2018; Yue et al. 2018), while viral proteins including AC11, AC51, AC66, AC75, AC78, GP41, AC93, P48, EXON0 and AC142 are required for nucleocapsid egress (Olszewski and Miller 1997; Fang et al. 2007; Ke et al. 2008; McCarthy et al. 2008; Yuan et al. 2008; Yuan et al. 2011; Tao et al. 2013; Tao et al. 2015; Guo et al. 2017a, b; Shi et al. 2017; Li et al. 2018; Qiu et al. 2019). Deletion of ac11, ac75, ac78, gp41, ac93, p48 or ac142 abrogated egress of nucleocapsids from the nucleus. By contrast, loss of ac51, ac66 or exon0 reduced the efficiency of nucleocapsid egress. A recent study showed that the nucleocapsids of BVs were ubiquitinated at much higher levels than those of ODVs, indicating that nucleocapsid ubiquitination (potentially catalyzed by the viral E3 ubiquitin ligase EXON0) may play a key role in nucleocapsid egress (Biswas et al. 2017). Exploring genes associated with nucleocapsid egress is important to elucidate the mechanism of nucleocapsid egress in baculoviruses.
ac13 (Genebank accession number KM667940.1), encoding a protein of 327 amino acids with a putative molecular mass of 38.7 kDa (Rohrmann 2019), is a conserved gene in all sequenced alphabaculoviruses. However, the function of ac13 in the viral life cycle remains unknown. To date, only a few studies have examined ac13 and its orthologs. Transcriptomic sequencing showed that ac13 was regulated by an early promoter and a late promoter (Chen et al. 2013). InterProScan (Jones et al. 2014) and NCBI Conserved Domain Search (Marchler-Bauer et al. 2017) analyses revealed that AC13 contained a DUF3627 protein domain of unknown function, which was conserved in all alphabaculovirus but not betabaculovirus orthologs. bm5, a homolog of ac13 in Bombyx mori NPV (BmNPV), was seemingly nonessential because the viral life cycle appeared normal when it was deleted (Ono et al. 2012). However, a recent study showed that although deletion of bm5 did not affect viral DNA replication, it decreased BV and OB production (Kokusho et al. 2016). In this study, we investigated the function of ac13 in the baculovirus life cycle. Temporal transcription and transcription initiation sites (TSSs) analyses showed that ac13, with an early promoter and a late promoter, was transcribed during the early and late stages of infection. However, the AC13 protein was only detected at the late infection and colocalized with the nuclear lamina. In addition, we determined the roles of ac13 in BV production and OB morphogenesis. Our results indicated that ac13 was not essential for viral genome replication, gene transcription, nucleocapsid assembly or OB formation. However, ac13 deletion reduced the efficiency of nucleocapsid egress and decreased the production of BVs. Thus, ac13 was essential for efficient nucleocapsid egress from the nucleus to the cytoplasm during BV production, but not for OB formation.
-
Sf9 cells (Invitrogen, Carlsbad, CA, USA) were cultured at 27 ℃ in Grace's medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Gibco, Grand Island, NY, USA). AcMNPV recombinant bacmids were constructed with the bacmid bMON14272 (Invitrogen) and maintained in E. coli strain DH10B (Invitrogen). Spodoptera exigua (S. exigua) larvae (Core Facility and Technical Support of Wuhan Institute of Virology, CAS, Wuhan, China) were reared on an artificial diet at 28 ℃.
The anti-AC13 polyclonal antiserum was prepared in rabbits according to previously published methods (Li et al. 2018). The polyclonal anti-GP64 and anti-VP39 antibodies were gifts from Prof. Zhihong Hu (Wuhan Institute of Virology, CAS, Wuhan, China). Mouse monoclonal antiactin and rabbit polyclonal anti-FLAG antibodies were purchased from Proteintech (Wuhan, China) and mouse monoclonal anti-lamin Dm0 antibody was purchased from the Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA, USA).
-
Sf9 cells (1.0 × 106 cells/well) were infected with AcMNPV at a multiplicity of infection (MOI) of 5 TCID50/cell and collected at 0, 3, 6, 12, 24, 36 and 48 h post-infection (h p.i.). Total RNA was extracted using RNAiso Plus (Takara, Toyoko, Japan) according to the manufacturer's instructions and quantitated using a Nanodrop-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Finally, transcripts were detected by PCR using gene-specific primer pairs (sequences were shown in Table S1) and the cDNA as a template.
For the time course analysis of AC13 expression, Sf9 cells (1.0 × 106 cells/well) were infected with AcMNPV at an MOI of 10 and harvested at 6, 12, 18, 24 and 48 h p.i. Subsequently, Western blotting was performed with antiAC13 (1:1000) as primary antibody and horseradish peroxidase-conjugated goat anti-rabbit (1:3000; Proteintech) as the secondary antibody. A protein ladder (Thermo Fisher Scientific) was used to judge protein sizes.
-
Sf9 cells (1.0 × 106 cells/well) were infected with AcMNPV at an MOI of 5 and collected at 4 and 24 h p.i. Total RNA was isolated using RNAiso Plus (Takara). The rapid amplification of 5' cDNA ends (5' RACE) reaction was performed with an ac13-specific primer (ac13-GSP1, sequence was shown in Supplementary Table S1) using a SMARTerⓇ RACE 5'/3' Kit (Takara) according to the manufacturer's protocol. The PCR products were cloned into the pRACE vector and sequenced.
-
An ac13 knockout bacmid was constructed as previously described (Datsenko and Wanner 2000; Li et al. 2015). First, a 618-bp sequence upstream and a 686-bp sequence downstream of the ac13 open reading frame (ORF) were amplified by PCR using the primer pairs ac13-US-F/R and ac13-DS-F/R (sequences were shown in Supplementary Table S1), respectively, and the AcMNPV bacmid as a template. A 1,137-bp fragment was amplified using the primer pairs CmR-F/R (sequences were shown in Supplementary Table S1) using the plasmid pUC18-CmR as the template. Subsequently, the 618-bp upstream fragment, the 686-bp downstream fragment and the 1137-bp fragment were double-digested with SacI/BamHI, HindIII/XhoI and BamHI/HindIII, respectively. The three restriction digestion fragments were gel purified and consecutively ligated into the pBlueScript II SK (+) vector to generate pSKac13US-CmR-ac13DS. A fragment, amplified using the primer pair ac13-US-F/ac13-DS-R and template pSKac13US-CmR-ac13DS, was used to electroporate E. coli BW25113 cells (containing bMON14727 and pKD46) to replace the N-terminal 146-bp (10,298 to 10,443 nt of the AcMNPV genome) of ac13 with the CmR cassette via λ Red homologous recombination. The resulting ac13-null bacmid, confirmed by PCR and DNA sequencing, was named bAcac13KO.
Subsequently, the polh and egfp genes were separately cloned into the pFastBacDual vector (Invitrogen) under the control of the polh and p10 gene promoters via restriction digestion and ligation to generate pFBD-ph-egfp. DH10B competent cells, containing a helper plasmid pMON7124 and the bacmid bAcac13KO, were transformed with the plasmid pFBD-ph-egfp to generate an ac13-null bacmid (bAcac13KO-ph) by Tn7-mediated transposition. Similarly, a wild-type control bacmid (bAc-ph) was generated by inserting the polh and egfp genes into the polh locus of bMON1427. To construct an ac13 rescue bacmid, a 1458-bp fragment containing the ac13 native promoter and ORF was amplified by PCR with the primer pair Dualac13-F1/R1 (sequences were shown in Supplementary Table S1) from the bMON1427 template. The 1458-bp fragment was inserted in the plasmid pFBD-ph-egfp to produce pFBD-ph-ac13-egfp via homologous recombination. This vector was then used to transform DH10B competent cells (containing bAcac13KO and pMON7124) to generate an ac13-rescue bacmid (bAcac13REP-ph). Meanwhile, another ac13 rescue bacmid bAcac13FlagREP-ph (Supplementary Fig. S1), encoding a FLAG tag at its 3'- end, was constructed using the same method. All recombinant bacmids were confirmed by PCR.
-
The transient expression plasmid pIB-egfp was constructed using FastCloning (Li et al. 2011). Briefly, the pIB/V5-His vector (Invitrogen) and insert egfp fragment were amplified by PCR (prime sequences were shown in Supplementary Table S1). The egfp fragment 16 bp sequence was homologous with the vector. The PCR products were digested with DpnI (Takara) at 37 ℃ for 1 h, and then used to transform E. coli DH5α competent cells. Subsequently, the ac13 ORF was amplified from the AcMNPV genome and subcloned into pIB-egfp in-frame with the egfp fragment to generate pIB-ac13egfp via the same method. Based on the pIB-ac13egfp vector, the pIB-ac13ΔNLSegfp vector bearing a truncated ac13 gene with an NLS deletion (aa 260–270) was also constructed by FastCloning.
-
Sf9 cells were seeded in the six-well plate (1.0 × 106 cells/ well) and allowed to attach for 2 h at 27 ℃. The cells were transfected in triplicate with 10 μg of each bacmid DNA (bAcac13KO-ph, bAcac13REP-ph and bAc-ph) using 8 μL of Cellfectin II (Invitrogen) according to the manufacturer's instructions. The transfection buffer was then replaced with fresh Grace's medium after incubation for 5 h. The BVs contained in the supernatant were called vAcac13KO-ph, vAcac13REP-ph or vAc-ph. The supernatants were harvested at 24, 48, 72, 96 and 120 h p.t. and viral titers were determined using the endpoint dilution assay (Lei et al. 2020). Cells were infected in triplicate with each virus at an MOI of 0.002. After viral absorption for 1 h at 27 ℃, the infection mixture was replaced with fresh Grace's medium, and the time point was designated 0 h p.i. The supernatants were harvested at 12, 24, 48, 72, 96 and 120 h p.i., and viral titers were determined using the endpoint dilution assay (Lei et al. 2020). Statistical analysis was performed using one-way analysis of variance (ANOVA).
-
The qPCR analysis was performed as previously described (Vanarsdall et al. 2005) with some modifications. Briefly, Sf9 cells (1.0 × 106 cells/well) were transfected with bAcac13KO-ph or bAc-ph in triplicate and harvested at 0, 12 and 24 h p.t., respectively. Total DNA was extracted using a Universal Genomic DNA extraction kit version 5.0 (Takara) and digested with DpnI (Takara) at 37 ℃ overnight. Finally, qPCR was performed with the gp41-specific primer pairs (sequences were shown in Table S1) by a CFX96 real-time system (Bio-Rad). The reverse transcription RT-qPCR analysis was performed as previously described (Peng et al. 2012) with some modifications. Sf9 cells (1.0 × 106 cells/well) were transfected with bAcac13KO-ph or bAc-ph in triplicate and collected at 24 h p.t. Total RNA was isolated using RNAiso Plus (Takara), and total cDNA was obtained via reverse transcription using an iScript cDNA synthesis kit (Bio-Rad). Subsequently, qPCR was performed with five primer pairs of selected viral genes (sequences were shown in Supplementary Table S1) using a CFX96 real-time system (BioRad). The 18S rRNA was selected as the endogenous reference.
-
Confocal immunofluorescence microscopy was performed as previously described (Xu et al. 2019). Sf9 cells were seeded (4 × 105 cells/dish) in a glass dish and allowed to attach for 2 h, then infected with vAcac13FlagREP-ph at an MOI of 5. The cells were fixed with 4% paraformaldehyde for 10 min at the designated time points. After washing with phosphate-buffered saline (PBS), the fixed cells were treated with 0.2% (v/v) Triton X-100 for 10 min and then blocked with PBS containing 5% (w/v) bovine serum albumin and 0.1% (v/v) Tween-20 for 30 min. Subsequently, the cells were incubated with rabbit anti-FLAG polyclonal antibody (1:500) and mouse anti-lamin Dm0 monoclonal antibody (1:500) for 1 h followed by Alexa Fluor 594 goat anti-rabbit IgG (1:1000, Invitrogen) and Alexa Fluor 647 goat anti-mouse IgG (1:1000, Invitrogen) for 1 h in dark. Finally, the cell nuclei were stained with Hoechst 33,258 (Beyotime, Shanghai, China) for 7 min in the dark and examined using the PerkinElmer UltraView VOX system with a 60 × oil-immersion.
-
Transmission electron microscopy (TEM) analysis was performed as previously described (Qin et al. 2019). Sf9 cells were seeded (1 × 106 cells/well) and allowed to adhere for 2 h, then infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph. At 48 h p.i., the cells were fixed with 2.5% (v/v) glutaraldehyde for 2 h. Ultrathin sections were visualized using FEI Tecnai G2 20 TWIN transmission electron microscope. Twenty intact cells infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph were randomly chosen to analyze nucleocapsid egress. The numbers of intranuclear and egressed nucleocapsids in each cell section were counted using ImageJ software (https://imagej.nih.gov/ij/) and compared using the Kruskal–Wallis test followed by Dunn's multiple comparison test. For immunoelectron microscopy (IEM) analysis, Sf9 cells were infected with vAcac13FlagREP-ph at an MOI of 10. At 48 h p.i., the cells were fixed with 1% paraformaldehyde-0.5% glutaraldehyde for 10 min at 4 ℃ and refixed with 2% paraformaldehyde 2.5% glutaraldehyde for 1 h at 4 ℃, and then were dehydrated and embedded according to previously methods (Xu et al. 2019). Ultrathin sections were immunostained with anti-FLAG pAb (1:50) as the primary antibody. Goat anti-rabbit IgG coated with gold particles (10 nm; Sigma, Darmstadt, Germany) was used as the secondary antibody (1:50). The Ultrathin sections also were visualized by an FEI Tecnai G2 20 TWIN transmission electron microscope.
-
OBs were purified from larvae by differential centrifugation according to the method described by Gross et al. (Gross et al. 1994) and observed by SEM (Hitachi SU8010) and TEM (FEI Tecnai G2 20 TWIN) as described previously (Kuang et al. 2017). To observe ODVs embedded within OBs, 10 μL of OB suspension (108 OBs/mL) were loaded onto a copper grid for 10 min. Filter paper was used to remove the remaining solution from the grid. Then, 10 μL of dissolution buffer [0.1 mol/L Na2CO3, 0.17 mol/L NaCl, 0.01 mol/L EDTA (pH 10.5)] was added to dissolve the OBs for 1 min. After removing the dissolution buffer, the grid was stained with 2% (w/v) phosphotungstic acid (pH 5.7) for 1 min. The grids were kept at room temperature overnight and observed by TEM. The ODVs in each OB were counted using ImageJ software and their numbers were compared using the Kruskal–Wallis test followed by Dunn's multiple comparison test.
Cells, Viruses, Insects and Antibodies
Transcription and Expression Analysis
Transcription Initiation Sites Analysis
Construction of ac13 Knockout and Repaired Bacmids
Construction of ac13 Subcellular Localization Plasmids
Transfection and Infection Assays
Quantitative Analysis of Viral Genome Replication and Viral Gene Transcription
Fluorescence Confocal Microscopy Analysis
Electron Microscopy Analyses
Scanning Electron Microscopy (SEM), TEM and Negative Staining Analyses
-
Transcriptomic analysis showed that two different TSSs were located upstream of the ac13 translation initiation codon (Chen et al. 2013). Temporal transcription patterns showed that the product of ac13 was detected as early as 3 h p.i. and persisted up to 48 h p.i. (Fig. 1A). The 5' RACE analysis revealed that the TSSs mapped to the first G of the atypical baculovirus early promoter motif GCAGT, located 217 nt upstream of the ac13 ORF start codon, and the first A of the typical late promoter motif TAAG, located 56 nt upstream of the ac13 ORF start codon (Fig. 1B). These results indicated that ac13 gene was regulated by an early promoter and a late promoter, and it was transcribed during the early and late stages of infection.
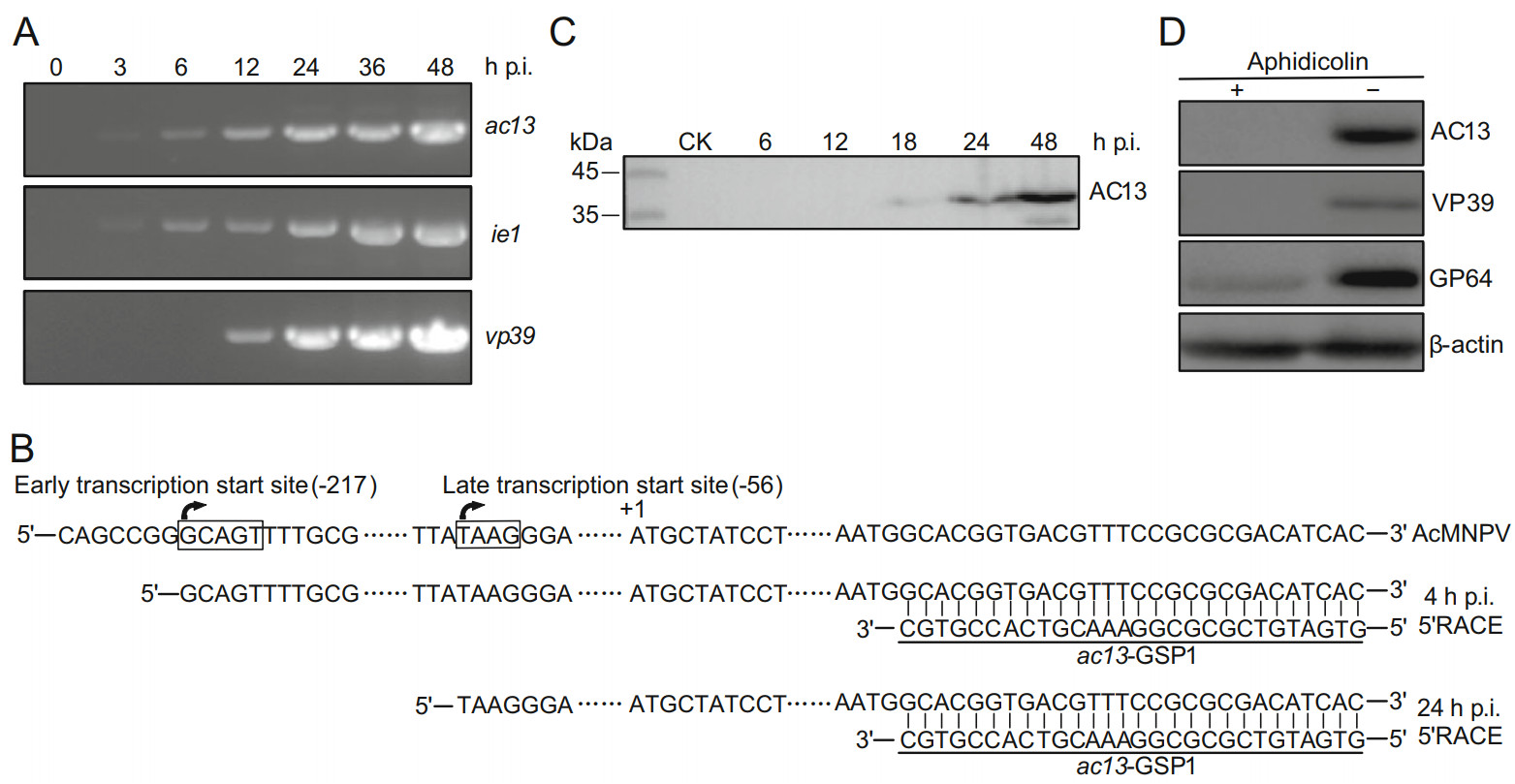
Figure 1. Transcription and expression analyses of ac13. A RT-PCR analysis of ac13 transcription. Total RNA was extracted from AcMNPV-infected cells at the indicated time points. The transcripts were amplified with ac13-, ie1- or vp39-specific primes, respectively. B 5' RACE analysis of ac13 TSS. Total RNA was extracted from AcMNPV-infected cells at 4 and 24 h p.i. and subjected to 5' RACE analysis. The late promoter (TAAG) and the early promoter (GCAGT) were denoted in box and the two TSSs were shown with arrowhead. C Western blot analysis the temporal expression of AC13. Sf9 cells, infected with AcMNPV at an MOI of 10, were harvested at the indicated time points and detected with anti-AC13 antibody. D Western blot analysis of the expression of AC13 with aphidicolin. The AcMNPV-infected cells were treated with 5 μg/mL aphidicolin (+) or DMSO control (-) at 0 h p.i. and then processed at 24 h p.i. and detected with anti-AC13. The VP39, GP64 and β-actin were used as controls.
The temporal expression profile of AC13 was determined by Western blotting of AcMNPV-infected cells at designated time points. A band of approximately 39 kDa, close to the predicted molecular mass of AC13, was detected from 18 to 48 h p.i. (Fig. 1C). To further determine whether AC13 was expressed during the late infection, AC13 was detected in AcMNPV-infected cells via the presence of aphidicolin, which inhibits viral DNA replication and thus prevents viral late gene expression. No AC13 expression was observed in aphidicolin-treated cells, while AC13 was detected in control cells treated with dimethyl sulfoxide (DMSO) at 24 h p.i. (Fig. 1D). Expression of VP39 was only detected in DMSO-treated cells, while expression of GP64 was detected both in aphidicolin-treated and DMSO-treated cells (Fig. 1D). These data showed that despite the transcription of ac13 could be detected during the early and late stages of infection, AC13 protein was only detected at the late stage. Thus, ac13 can be designated as a late viral gene.
-
To investigate the function of AC13 in the viral life cycle, its subcellular localization was analyzed by confocal microscopy. Sf9 cells, infected with vAcac13FlagREP-ph at an MOI of 5, were fixed at 12, 18, 24, 36, 48 and 72 h p.i. AC13 was detected by immunofluorescence using confocal microscopy. As shown in Fig. 2A, AC13 fluorescence was predominantly localized to the nuclear ring zone and mainly colocalized with the nuclear lamina from 12 until 72 h p.i. Immunoelectron microscopy was also used to assess the localization of AC13 in cells infected with vAcac13FlagREP-ph. At 48 h p.i., colloidal-gold-labeled AC13 was predominantly localized to the perinuclear and nuclear membranes (Fig. 2B). This result agreed with the immunofluorescence data.
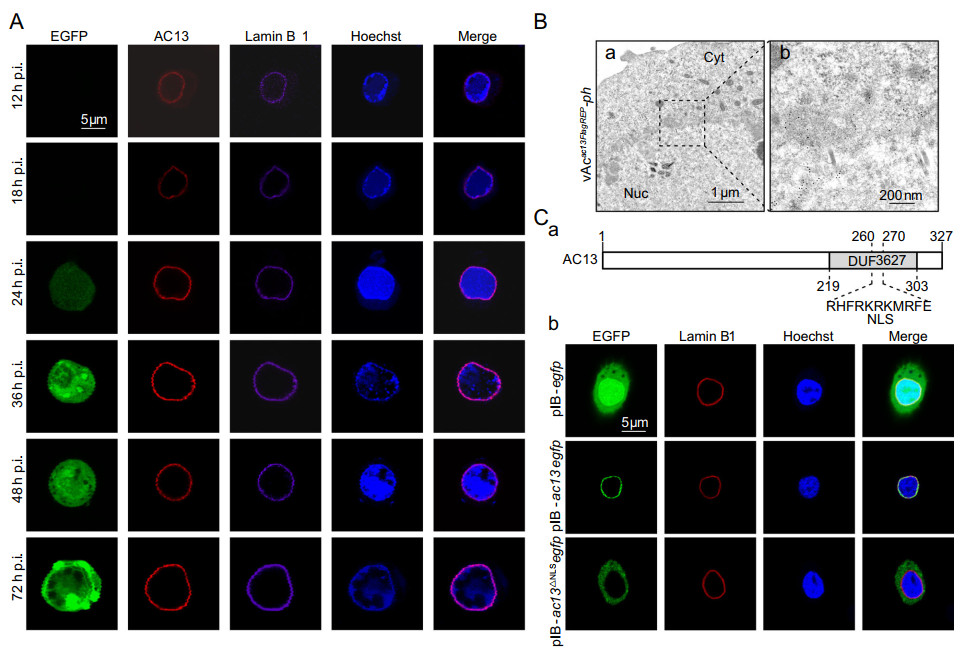
Figure 2. Subcellular localization analysis of AC13. A Immunofluorescence analysis. Sf9 cells, infected with vAcac13FlagREP-ph at an MOI of 5, were fixed with paraformaldehyde at the indicated time points and immunostained with an anti-FLAG antibody to detect AC13 (red), an anti-lamin Dm0 antibody to detect Lamin B1 (purple). EGFP was an indicator of cells infected with virus (green). The nuclei were stained with Hoechst33258 (blue). B Immunoelectron microscopy analysis. Sf9 cells were infected with vAcac13FlagREP-ph at an MOI of 10 and harvested at 48 h p.i. The ultrathin sections were probed with anti-FLAG antibody as the primary antibody and goat anti-rabbit IgG coated with gold particles (10 nm) as the secondary antibody. C-a Schematic representation of the NLS of AC13. The NLS of AC13 was predicted in the DUF3627. C-b Fluorescence microscope analysis. Sf9 cell, transfected with the plasmids pIB-ac13egfp, pIB-ac13ΔNLSegfp or pIB-egfp, were analyzed by immunofluorescence microscopy at 24 h p.t. EGFP was an indicator of AC13 (green), and an anti-lamin Dm0 antibody was used to detect the Lamin B1 in cells (red). The nuclei were stained with Hoechst 33,258 (blue).
Further bioinformatic analysis indicated that there was a putative nuclear localization signal (NLS) motif in the DUF3627 domain of AC13 (Fig. 2C-a). To examine the localization of AC13 in the absence of viral infection or the NLS motif, Sf9 cells were transfected with the pIB-ac13egfp, pIB-ac13ΔNLSegfp or pIB-egfp plasmids. As shown in Fig. 2C-b, pIB-ac13egfp fluorescence was detected at the nuclear lamina, whereas pIB-ac13ΔNLSegfp, encoding a disrupted NLS, fluorescence was observed only in the cytoplasm. As a control, pIB-egfp showed homogenous fluorescence throughout the cytoplasm and nucleus. These results indicated that AC13 was located to the nuclear lamina independently of viral infection, but nuclear import required the NLS motif.
-
To investigate the function of ac13 in the AcMNPV life cycle, an ac13-null bacmid, bAcac13KO-ph, an ac13-rescue bacmid, bAcac13REP-ph, and a pseudo-wild-type bacmid, bAc-ph (Fig. 3), were constructed. All bacmids were confirmed by PCR (primer sequences were shown in Supplementary Table S1).
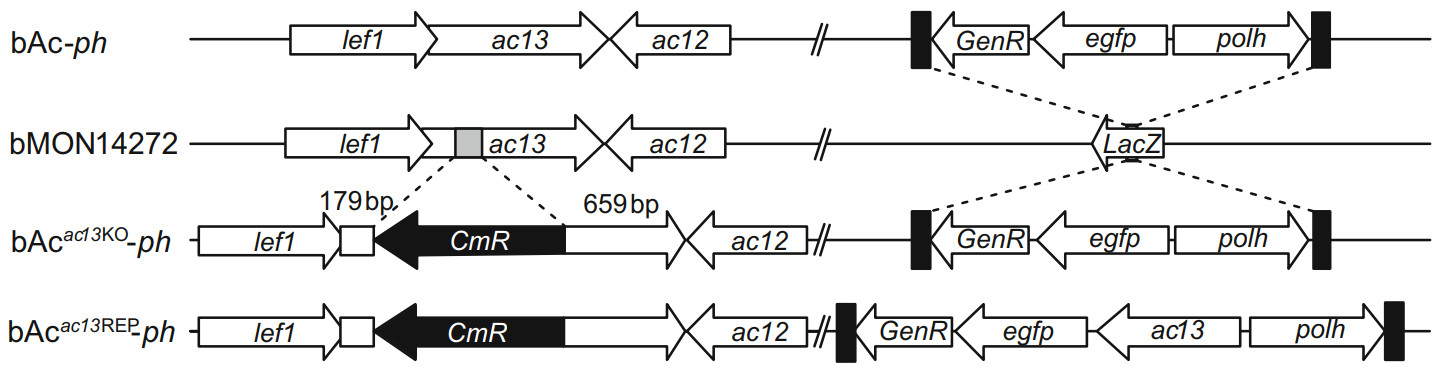
Figure 3. Schematic diagram of bAcac13KO-ph, bAcac13REP-ph and bAc-ph construction. Using the bacmid bMON14272, the bAcac13KO was generated by replacing 146 bp fragment of the ac13 ORF with a chloramphenicol resistance (CmR) gene cassette via homologous recombination. The egfp and polh genes were inserted into the polh locus of bAcac13KO via Tn7-mediated transposition to generate bAcac13KO-ph. The ac13 together with the egfp and polh genes were inserted into the polh locus of bAcac13KO to generate bAcac13REP-ph. bAc-ph was constructed by inserting the egfp and polh genes into the polh locus of bMON14272.
To determine the effects of the absence of ac13 on viral proliferation and OB morphogenesis, Sf9 cells were transfected with the bAcac13KO-ph, bAcac13REP-ph or bAc-ph bacmids and monitored under a fluorescent microscope. No obvious differences were found in the numbers of fluorescent cells among the three bacmids at 24 h p.t. (Fig. 4A, upper panel), indicating that the three bacmids had comparable transfection efficiencies. However, the number of fluorescent bAcac13KO-ph-transfected cells increased much less than those of bAcac13REP-ph and bAc-ph from 24 to 96 h p.t. (Fig. 4A, upper and middle panels). To further examine the effects of ac13 deletion on viral proliferation, we collected BV supernatants from cells transfected with each bacmid at the indicated time points. Virus titers were determined using the endpoint dilution assay, and performed one-step growth curve analysis. Viruses produced from the bAcac13REP-ph and bAc-ph bacmids showed similar growth kinetics, reaching 7.0 × 108 and 4.0 × 108 TCID50/ml at 96 h p.t., respectively. However, the viral titer of bAcac13KO-ph was reduced by 400-fold compared with bAcac13REP-ph or bAc-ph at 120 h p.t. (P < 0.001) (Fig. 4B). These results indicated that although bAcac13KO-ph-transfected cells were able to produce infectious BVs, the efficiency of BV production was impaired. Light microscopy analysis revealed that a large proportion of cells transfected with bAc-ph or bAcac13REP-ph contained OBs at 96 h p.t., whereas only a small proportion of bAcac13KO-ph-transfected cells contained OBs (Fig. 4A, lower panel). However, no obvious differences were found in the average numbers of OBs produced by each cell transfected with bAcac13KO-ph, bAcac13REP-ph or bAc-ph at 96 h p.t. (Fig. 4A, lower panel and Fig. 4C), indicating that ac13 deletion had no effect on OB formation.
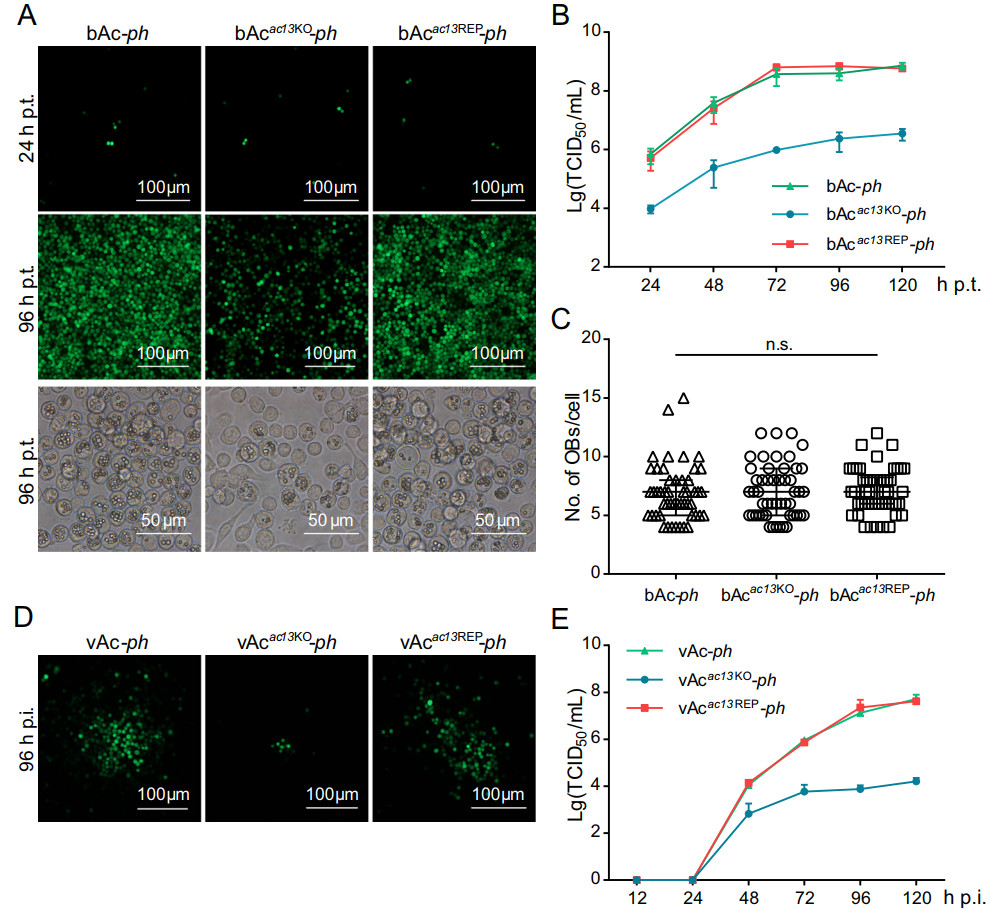
Figure 4. Analyses of viral replication and OB formation in the transfected/infected cells. A Fluorescence microscopy of cells transfected with the bacmids of bAc-ph, bAcac13KO-ph or bAcac13REP-ph at 24 or 96 h p.t. (upper and middle panels). Light microscopy of cells transfected with each bacmid at 96 h p.t. (lower panel). B Onestep growth curve generated from bAc-ph-, bAcac13KO-ph- or bAcac13REP-ph-transfected cells. The supernatants of Sf9 cells, transfected with bAc-ph, bAcac13KO-ph or bAcac13REP-ph, were harvested at the designated time points and quantified by the endpoint dilution assay. Each data points represent average titers from three separate transfections. Error bars represent standard deviations (SD). C OB production in each cell. The number of envisaged-visible OBs was under phase contrast microscope counted at 96 h p.t., and more than 50 cells were counted for each condition. n.s. indicates no significance, P > 0.05. D Fluorescence microscopy images of Sf9 cells infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph at an MOI of 0.002 at 96 h p.i. E One-step growth curve generated from vAc-ph-, vAcac13KO-ph- or vAcac13REP-ph-infected cells. Sf9 cells were infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph at an MOI of 0.002. The supernatants were collected at the indicated time points and determined by the endpoint dilution assay. Each data points represent average titers from three separate infections. Error bars represent SD.
To confirm the results obtained following bacmid transfection, Sf9 cells were infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph at an MOI of 0.002. The supernatants were collected and BV titles were determined by the endpoint dilution assay at the designated time points, and performed one-step growth curve analysis. As shown in Fig. 4D and 4E, vAcac13REP-ph showed similar growth kinetics to vAc-ph. However, BV production in vAcac13KO-ph-infected cells was significantly reduced compared with that in cells infected with vAc-ph and vAcac13REP-ph. Taken together, these data suggested that ac13 deletion significantly decreased infectious BV production, but did not affect OB formation in cells.
-
The BV life cycle includes replication of viral genome, assembly of progeny nucleocapsids, egress from the nucleus, transport through the cytoplasm, and budding from the plasma membrane where the BV gains its envelope. To determine whether reduced BV production in vAcac13KO-ph-infected cells resulted from a defect in viral genome replication, viral genome replication was compared between bAcac13KO-ph- and bAc-ph-transfected cells by quantitative PCR (qPCR) within 24 h p.t., before secondary infection by BVs could occur (Qiu et al. 2019). Sf9 cells were transfected with equal amounts of bAcac13KO-ph or bAc-ph bacmid DNA and collected at 0, 12 and 24 h p.t. Total intracellular DNA was extracted and treated with DpnI to eliminate input bacmid DNA. The viral genome copy number was measured by qPCR using gp41-specific primers (sequences were shown in Table S1) as previously reported (Vanarsdall et al. 2005). As shown in Fig. 5A, the viral DNA content in bAcac13KO-ph and bAc-ph-transfected cells both increased with similar rates from 0 to 24 h p.t. (P > 0.05), indicating that ac13 deletion did not affect viral genome replication. Subsequently, the transcription of five viral genes, including two early genes (ie1 and pe38), one early-late gene (gp64) and two late genes (vp39 and polh), was analyzed by RT-qPCR. There were not significant differences in the transcript levels of any genes between bAcac13KO-ph- and bAc-ph-transfected cells (P > 0.05) (Fig. 5B), suggesting that ac13 deletion did not affect early or late viral gene transcription. These data revealed that ac13 deletion did not impair viral genome replication or gene transcription.
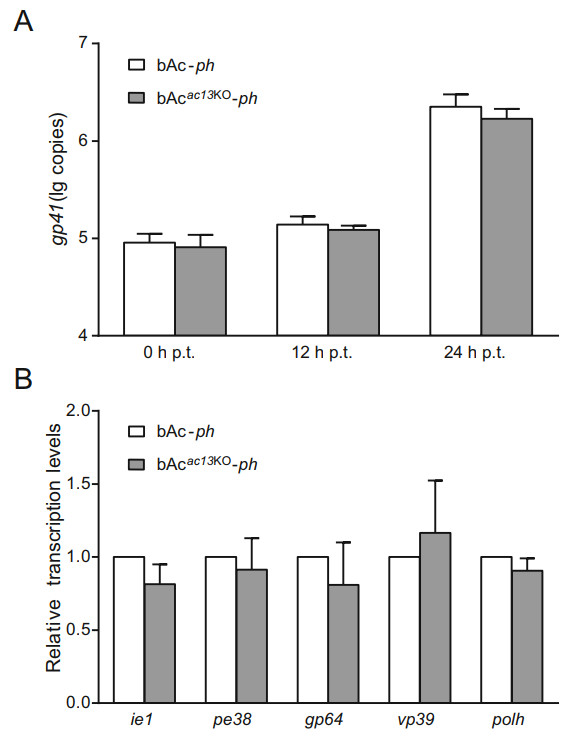
Figure 5. Analyses of viral genome replication and viral genes transcription. A qPCR analysis of viral DNA replication. Total cellular DNA was extracted at the indicated time points from cells, transfected with the bacmids of bAcac13KO-ph or bAc-ph, and digested with restriction enzyme DpnI to eliminate input bacmid DNA. The genomes copies were analyzed with qPCR using the gp41-specific primer pairs. The values represent the averages from three independent assays. Error bars indicate SD. B RT-qPCR analysis of viral genes transcription at 24 h p.t. Total cellular RNA was extracted at 24 h p.t. from cells which were transfected with bAcac13KO-ph or bAc-ph. The transcription of selected viral genes was measured with RT-qPCR. The transcript level of each gene was normalized to that of the cell 18S rRNA. The values represent the averages from three independent assays, and error bars represent SD.
-
To further explore the impediments to BV production in the absence of ac13, TEM was used to examine thin sections generated from cells infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph at an MOI of 10 at 48 h p.i. As shown in Fig. 6A, the typical symptoms of baculovirus infection appeared both in vAcac13KO-ph- and vAc-ph-infected cells. These included an enlarged nucleus with a net-shaped virogenic stroma, a large number of rod-shaped electron-dense nucleocapsids within the virogenic stroma, and mature ODVs with multiple nucleocapsids and ODVcontaining OBs around the ring zone. As expected, vAcac13REP-ph-infected cells exhibited similar characteristics to those of cells infected with vAc-ph. According to the results, ac13 deletion did not affect either nucleocapsid assembly or OB formation.
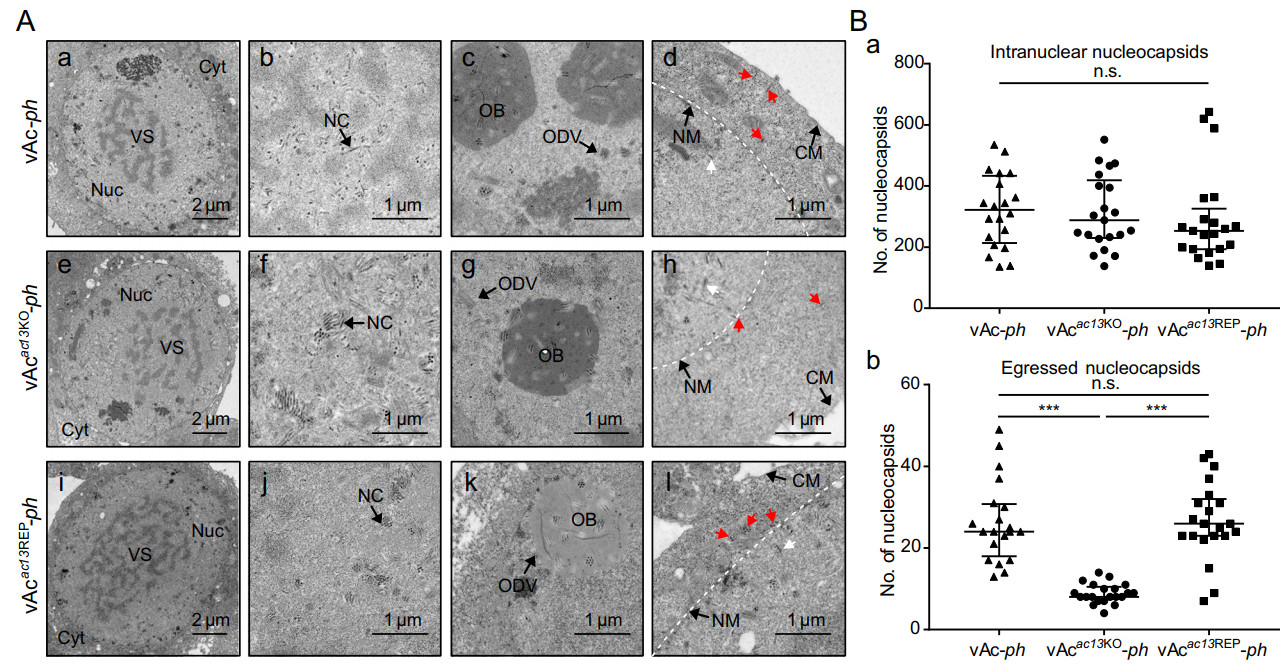
Figure 6. Transmission electron microscopy analyses of cells infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph. A Sf9 cells, infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph at an MOI of 10, were fixed at 48 h p.i. and prepared for TEM. (A-a to A-d) Cells infected with vAc-ph. (A-e to A-h) Cells infected with vAcac13KO-ph. (A-i to A-l) Cells infected with vAcac13REP-ph. (A-a, A-e and A-i) Enlarged nucleus (Nuc) and virogenic stroma (VS) in cells infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph. (A-b, A-f and A-j) Normal electron-dense nucleocapsids (NC) in cells infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph. (A-c, A-g and A-k) ODVs containing multiple nucleocapsids and OBs embeding normal ODVs in cells infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph. (A-d, A-h and A-l) Nucleocapsids residing in the cytoplasm or budding from the nuclear or cytoplasmic membranes were indicated with red arrows, while nucleocapsids residing in the nucleus were indicated with white arrows in cells. The nuclear membrane was shown with white dotted line. Cyt, cytoplasm; NM, nuclear membrane; CM, cytoplasmic membrane. B The numbers of intranuclear nucleocapsids (n.s. indicates no significance, P > 0.05) (B-a) and egressed nucleocapsids (*** indicates P < 0.001, n.s. indicates no significance, P > 0.05) (B-b) were determined. Numbers were calculated from 20 cells.
AC13 is localized to the nuclear lamina, and therefore it might play a role in egress of nucleocapsids from the nucleus. The TEM analysis was used to assess whether ac13 deletion had any effect on nuclear egress. According to methods previously reported (Fang et al. 2007; Ke et al. 2008; Qiu et al. 2019), we counted and compared the numbers of intranuclear and egressed nucleocapsids in 20 randomly chosen cells infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph. The egressed nucleocapsids included nucleocapsids exiting the nuclear membrane, in transport through the cytoplasm and budding from the cytoplasmic membrane (Fig. 6A-d, 6A-h and 6A-i). Intranuclear nucleocapsids of vAcac13KO-ph-infected cells were comparable to those of vAcac13REP-ph- and vAc-ph-infected cells (Fig. 6B-a). By contrast, the numbers of egressed nucleocapsids in vAcac13KO-ph-infected cells were reduced by 3.0 folds compared with those of vAcac13REP-ph- or vAc-ph-infected cells (P < 0.001) (Fig. 6B-b). Taken above results together, ac13 deletion impaired the efficiency of nucleocapsid egress from the nucleus to the cytoplasm.
-
According to the above results, ac13 deletion did not affect the number of OBs formed within each cell. To further investigate whether ac13 deletion had an effect on OB morphogenesis in larvae, SEM, TEM and negative staining analyses were performed on OBs purified from S. exigua cadavers infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph. As shown in Fig. 7A, the OBs formed within vAcac13KO-ph-infected larvae had smooth surfaces and sharp edges and contained normal ODVs, similar to those from vAc-ph- or vAcac13REP-ph-infected larvae. Subsequently, we dissolved, negatively stained and imaged the OBs of vAc-ph, vAcac13KO-ph or vAcac13REP-ph by TEM. And we counted the numbers of ODVs within OBs of the three viruses using ImageJ software. The average number of ODVs per OB of vAcac13KO-ph was comparable to those of vAc-ph or vAcac13REP-ph (P > 0.05) (Fig. 7B). Taken together, these results showed that ac13 deletion did not affect OB morphogenesis in S. exigua larvae.

Figure 7. SEM, TEM and negative staining analyses of OBs. A OBs, purified from S. exigua larvae infected with vAc-ph, vAcac13KO-ph or vAcac13REP-ph, were observed with SEM (upper panel), TEM (middle panel) and negative staining after treating with dissolution buffer on the grid (lower panel). B Numbers of ODVs embedded in each OB. More than 40 OBs of each virus were analyzed. n.s. indicates no significance (P > 0.05).
ac13 Is a Late Viral Gene
AC13 Is Predominantly Localized to the Nuclear Lamina
ac13 Is Essential for BV Production but Not for OB Formation
ac13 Is Not Required for Replication of Viral Genome or Transcription of Viral Genes
ac13 Is Required for Efficient Nuclear Egress of Nucleocapsids
ac13 Is Not Required for OB Morphogenesis in Larvae
-
The ac13 gene is conserved in all sequenced alphabaculoviruses, implying that it may play an important role in the viral life cycle. In this study, we investigated the role of ac13 in AcMNPV by constructing an ac13-null bacmid (bAcac13KO-ph). We determined that ac13 was required for efficient egress of nucleocapsids from the nucleus to the cytoplasm, but not for OB formation.
The 5' RACE analysis indicated that ac13 was regulated by an atypical early promoter (GCAGT) and a canonical late promoter (TAAG) in AcMNPV-infected Sf9 cells. The TSS of the late promoter was consistent with previous studies. However, the TSS of the early promoter differed by 2 nt from previous data generated from AcMNPVinfected T. ni cells. A potential explanation for this discrepancy might be that different hosts impact the TSS usage of AcMNPV. Temporal transcription analysis revealed that ac13 was transcribed from 3 h p.i. and persistent up to 48 h p.i. Thus, ac13 might play a role at the early and late stages of infection, although the AC13 protein was only detected during the late stage. However, we cannot rule out the possibility that the levels of AC13 expression during the early stage were too low to be detected. It is also possible that ac13 may play a role in early infection not through an encoded protein but other types of gene products (i.e., peptides or non-coding RNAs), or may participate as a DNA element. For example, ac83 is involved in nucleocapsid envelopment via an internal cisacting element (Huang et al. 2017).
Subcellular localization analysis showed that AC13 localized at the nuclear lamina independent of viral infection. Protein nuclear import typically requires an NLS, which assists transport through the nuclear pore complex into the nucleus. Previous studies identified several proteins with NLSs in AcMNPV including LEF-3, DNApol, IE1 and BV/ODV-C42 (Olson et al. 2002; Victoria et al. 2009; Feng and Krell 2014). Bioinformatic and confocal microscopy analyses indicated the presence of an NLS motif in AC13, which played an essential role in its nuclear import (Fig. 2C). However, the bioinformatic analysis suggested that AC13 is not an integral membrane protein. Yet-unknown proteins may facilitate the nuclear membrane accumulation of AC13.
It was reported that bm5 encoded a multifunctional protein that regulates viral transcription and OB formation and contributed to BV production (Kokusho et al. 2016). However, in our study, observation of cells transfected with bAcac13KO-ph or bAc-ph demonstrated that ac13 deletion reduced BV production by 99.7%, but did not affect OB formation. The CmR cassette was reversibly inserted in the ac13 ORF, completely disrupting ac13 expression without affecting transcription or expression of the neighbor genes lef1 and ac12. The phenotype of the ac13 knockout could be rescued by a repair virus. This discrepancy may result from different measurement methods use. In Kokusho et al. (2016) study, OB formation was confirmed by counting the number of OBs in the whole dish. As bm5 deletion reduced BV production and decreased the number of cells undergoing secondary infection, the number of bm5-null-infected cells may have been lower than the number of cells infected with wild-type virus. In our study, we assessed OB formation by counting the number of OBs per cell and observed the shape and inner structure of OBs via SEM and TEM. In addition, the differences may also have resulted from different viruses. Several genes have been reported to show discrepancies in knockout phenotypes between AcMNPV and BmNPV. For instance, gp41 is essential for AcMNPV replication (Li et al. 2018) but not for BmNPV replication (Ono et al. 2012). Additionally, ac51 is required for BV production but not for OB morphogenesis in AcMNPV (Qiu et al. 2019), while bm40, an orthologous gene of ac51 in BmNPV, is essential for BV and OB morphogenesis (Shen et al. 2018).
The nuclear membrane accumulation of AC13 suggested that it may be involved in nucleocapsid egress from the nucleus. In the alphaherpesviruses, pUL31 and pUL34, which are required for egress of nucleocapsids from the nucleus to the cytoplasm, are co-dependently localized to the nuclear rim (Klupp et al. 2007). Largely based on the results of TEM analysis, nucleocapsid egress from the nucleus is thought to occur through a process of budding from the nuclear membrane (Blissard and Theilmann 2018), and a recent study demonstrated that baculovirus nucleocapsid egress from the nucleus by disrupting the nuclear membrane (Ohkawa and Welch 2018). We used TEM to examine and compare cells infected with vAcac13KO-ph, vAcac13REP-ph or vAc-ph. The results showed that ac13 deletion did not affect nucleocapsid assembly or progression of viral infection into very late phases to form OBs, but impaired efficient egress of nucleocapsids from the nucleus to the cytoplasm. Several AcMNPV genes have been reported to be involved in nuclear egress of nucleocapsids including ac11, ac51, ac66, ac75, ac78, gp41, ac93, p48, exon0 and ac142. The first to be identified, exon0, is not strictly essential for the production of BVs because a few nucleocapsids in cells infected with an exon0-null virus did pass through the nuclear membrane. Recently, Qiu et al. demonstrated that the ac51 deletion affected the efficiency of nucleocapsid egress, but did not affect nucleocapsid assembly and ODV envelopment (Qiu et al. 2019). In this study, the genes required for the nuclear egress of nucleocapsids and production of BVs seemed to be divided into two categories (Qiu et al. 2019): (1) genes whose deletion did not affect nucleocapsid assembly but prevented nuclear egress of nucleocapsids, thus abrogating BV production and also interrupting ODV formation, and (2) genes whose deletion did not affect nucleocapsid assembly but decreased the efficiency of nuclear egress of nucleocapsids, thus decreasing BV production. According to previous reports, exon0, ac66 and ac51 belong to the second category. Our results confirm that ac13 is a fourth gene belonging to the second category.
Egress of baculovirus nucleocapsids from the nucleus is an essential process for morphogenesis of mature BVs, which is required to spread infection within susceptible cells and tissues. In this study, we firstly found that ac13 was required for efficient nuclear egress of nucleocapsids during BV production. The results may contribute to a better understanding of nucleocapsid egress in baculoviruses.
-
This work was supported by the National Key Research and Development Program of China (2017YFD0201206) and the WIV "One-Three-Five" strategic program (Y602111SA1 to XS). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Additionally, we thank Prof. Zhihong Hu from Wuhan Institute of Virology, CAS, for polyclonal antibodies (anti-GP64 and anti-VP39). We also thank Youling Zhu from the Core Facility and Technique Support, Wuhan Institute of Virology, for assistance with anti-AC13 antibody preparation. We thank the Core Facility and Technical Support of Wuhan Institute of Virology, CAS, for their technical assistance in fluorescence microscopy (Ding Gao and Juan Min) and electron microscope (Pei Zhang, Anna Du and Bichao Xu).
-
XC, XS and JH designed the experiments. XC, XY and CL carried out the experiments and analyzed the data. XC, XS and JH wrote the manuscript. XC, FQ, XS and JH checked and finalized the manuscript. All authors read and approved the final manuscript.
-
The authors declare that there are no conflicts of interest.
-
The animal experiment was approved by Institutional Review Board, Wuhan Institute of Virology, Chinese Academy of Sciences (Approval Number WIVA13201401).
















 DownLoad:
DownLoad: