-
Dear Editor,
Since the outbreak of COVID-19 at the end of 2019, its causative agent SARS-CoV-2 has been spreading around the world for one and half a year. During the long global circulation of SARS-CoV-2, mutations in the viral genome gradually emerged and accumulated, which have led to the emergence of Variants of Concerns (VOC). The mutation linage B.1.351 (also termed as 501Y.V2 or Beta variant by WHO) was announced in December 2020 and rapidly became the predominant lineage in South Africa. The B.1.351 is characterized by eight mutations in S protein, including K417N, E484K and N501Y in RBD (Tegally et al. 2021). The N501Y and E484K mutations have been demonstrated reducing neutralizing titer of antibodies, vaccine-induced and SARS-CoV-2-infected sera in vitro, which raised concerns of current vaccine efficacy against the mutation lineages (Li et al. 2021; Wang et al. 2021).
Previously, we have demonstrated the ACE2 humanized mice (hACE2), whose ACE2 (mACE2) genes were replaced with the human ACE2 by using CRISPR/Cas9 knock-in technology, are highly susceptive to SARS-CoV-2 isolated in early 2020 (Singh et al. 2020; Sun et al. 2020). Thereafter, the hACE2 mice model has been widely used to investigate the pathogenic mechanism and evaluate antivirals against SARS-CoV-2 (Lv et al. 2020; Ye et al. 2020; Zhu et al. 2020). However, the infectivity and pathogenicity of B.1.351 variant remains not determined.
Herein, we characterized the replication dynamic and clinical outcome of B.1.351 variant in hACE2 mice with the same protocol as previously described. Accordingly, two groups of 12-month-old female hACE2 mice were intranasally challenged with the B.1.351 variant at a dose of 1.2 × 104 plaque forming unit (pfu) per mouse for daily monitoring (n = 8) and tissue collection (n = 12), respectively (Fig. 1A). The 501Y.V2-infected mice began to show ruffled fur and reduced activity on 3 days post infection. Meanwhile, B.1.351 variant infection caused significant weight loss, and the average weight loss reached ~ 13% on 3 days post infection (Fig. 1B). Then the mice began to recover and no death occurred during the seven-day observation period. These results indicate that the B.1.351 variant is pathogenic for the hACE2 mice, and leads more severe clinical symptoms than the prototype strain (IME-BJ05), which led only ~ 10% weight loss and no obvious clinical symptoms (Sun et al. 2020).
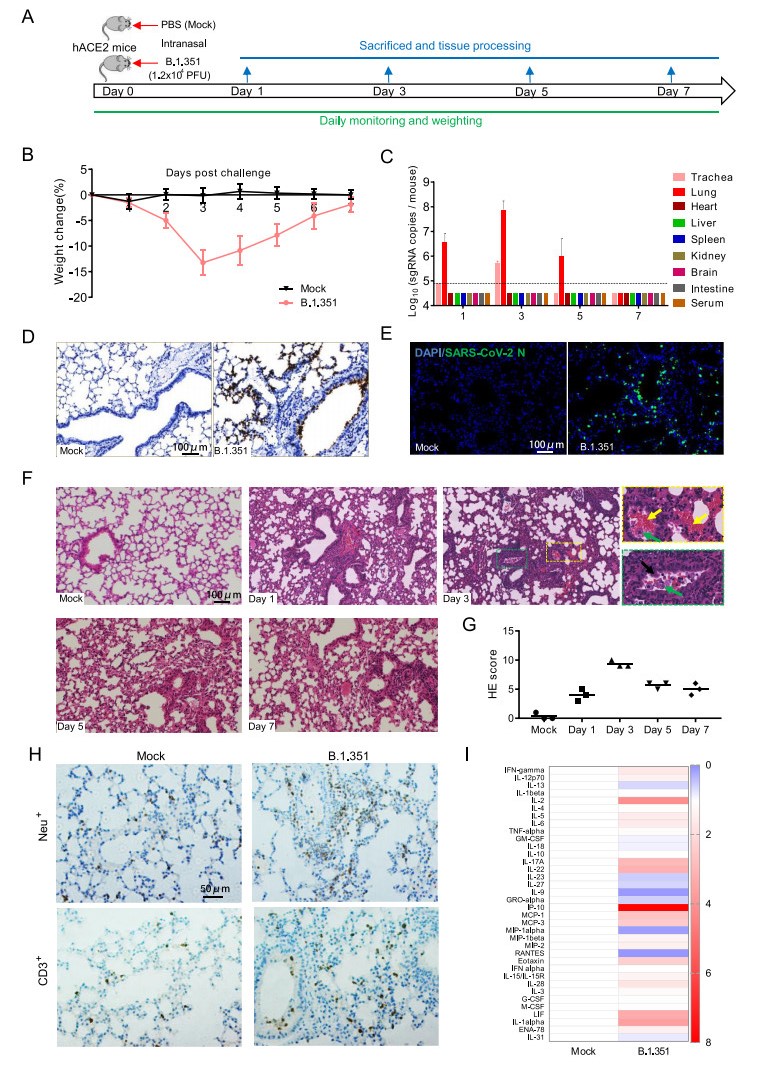
Figure 1. Infection of SARS-CoV-2 variant B.1.351 in hACE2 mice. A 12-month-old female hACE2 mice were intranasally infected with 1.2 × 104 pfu of SARS-CoV-2 variant B.1.351 or PBS (mock), and sacrificed on 1, 3, 5 and 7 dpi for tissue collection (n = 3). B Body weight changes of hACE2 mice intranasally infected with B.1.351 or PBS (mock) (n = 8). C Tissue distribution of SARS-CoV-2 sgRNA. Each tissue or serum was processed and viral sgRNA copies were analyzed by real-time qPCR. Dotted lines denote the detection limit. D ISH assay for SARS-CoV-2 RNA in lung tissues. Positive signals are shown in brown. E Immunostaining of lung tissues with a SARS-CoV-2 N protein-specific mAb. F Hematoxylin and eosin (H & E) staining analysis showing alveolar septal thickening, alveolar damage, denatured bronchiolar epithelial cells (black arrow), hemorrhage (yellow arrow), and macrophages infiltration (green arrow) in infected mice. G Semiquantitative analysis of the H & E-stained lung sections (n = 3). H IHC staining analysis for neutrophils (Neu+) and T cells (CD3+) in hACE2 mice on 3 days post infection. I Relative cytokine production in serum of infected hACE2 mice on 3 days post infection. For a more detailed description of methods used, please see the online Supplementary materials.
SARS-CoV-2 subgenomic RNA (sgRNA) quantitation was further performed for detecting the viral loads in the main tissues and serum of mice on day 1, 3, 5 and 7 post infection. Results showed that high level of SARS-CoV-2 sgRNA was detected in the lungs and tracheas from B.1.351-infected mice (Fig. 1C). An obvious sgRNA increasing trend was observed in lung tissues and tracheas during 1–3 days post infection, and then the sgRNA loads began to decrease (Fig. 1C). No viral sgRNA was detected in serum and other tissues. In situ hybridization (ISH) assay by RNAScope for the lung sections showed that the viral RNA mainly located in bronchiolar epithelial cells and at the alveolus walls (Fig. 1D). Immunostaining of lung sections also showed that the SARS-CoV-2 N protein expressed in both bronchiolar epithelial cells and alveolar cells (Fig. 1E). Taken together, these results indicate that the B.1.351 variant can effectively infect hACE2 mice, with robust viral replication in the respiratory system.
More importantly, histopathological observation showed that lung tissues from B.1.351-infected mice began to show injury as early as on day 1 post infection and became more severe on day 3 (Fig. 1F-G), characterized with denatured and collapsed bronchiolar epithelial cells (black arrow), thickened alveolar septa, alveolar damage, hemorrhage (yellow arrow), and macrophages infiltration (green arrow). Immunohistochemistry (IHC) staining indicated that the infiltrated inflammatory cells also include Neu+ neutrophils and CD3+ T cells (Fig. 1H), as well as the macrophages. The degree of alveolar septa thickening, alveolar and bronchiolar epithelial damage became lower, and the macrophages infiltration and hemorrhage were less on day 5 and 7 post infection compared with that on day 3 post infection, indicating a recovery process. Furthermore, Luminex cytokine analysis showed that SARS-CoV-2 infection led to elevated cytokine production in the sera, mainly including IL-1alpha, IL-2, IL-5, IL-6, IL-17A, IL-28, IL-22, IP-10, MCP-1, MCP-3, Eotaxin, and IFN-gamma (Fig. 1I). These results indicate that the B.1.351 infection can lead hACE2 mice to produce similar pulmonary pathology features as those observed in COVID-19 patients.
In conclusion, our present study confirmed that the SARS-CoV-2 variant B.1.351 can also effectively infect the hACE2 mice we generated previously, and lead to obvious pulmonary pathology. Notably, compared with the prototype strain IME-BJ05 we reported previously (4 × 105 pfu per mouse), the variant B.1.351 infection led more obvious clinical symptoms and weight loss at a much lower dose (1.2 × 104 pfu per mouse), indicating the increased virulence of the variant B.1.351 for hACE2 mice (Sun et al. 2020). Previous studies demonstrated that the N501Y and K417N mutations of SARS-CoV-2 S protein enhance the binding affinity to human ACE2 (Fratev 2020; Starr et al. 2020; Liu et al. 2021), which may contribute, at least partially, to the higher virulence of the variant B.1.351 for hACE2 mice. The variant B.1.351 effectively infects hACE2 mice and replicates in its respiratory system, and mainly distributes in bronchiolar epithelial cells and alveolar cells, which is similar to the prototype strain IME-BJ05 (Sun et al. 2020). Interestingly, in this study, no SARS-CoV-2 RNA (data not shown) or sgRNA was detected in the brain of mice infected with B.1.351, while high level of SARS-CoV-2 RNA in the brain of mice infected with IME-BJ05 was detected in our previous study. Whether the amino acid mutations in the mutation lineage affect this infection characteristic or which one plays the important role in the process needs to be further investigated.
In summary, our results showed that the hACE2 mice are susceptible to intranasal infection of the SARS-CoV-2 variant B.1.351, and robust viral replication was detected in respiratory system. Especially, the pathological changes observed in the hACE2 mice closely resemble those observed in COVID-19 patients. Thus, using the variant B.1.351 to infect the hACE2 mice described in our manuscript provides a small animal model for studying the transmission, pathogenesis, as well as understanding the unexpected clinical manifestations of the mutation lineage. This model will also be valuable for testing vaccines and therapeutics to combat the variant B.1.351.
HTML
-
We acknowledge NPRC for sharing the 501Y.V2 variant. This work was supported by the National Key Research and Development Project of China (2020YFC0842200, 2020YFA0707801, and 2021YFC0863300) and the National Natural Science Foundation of China (No. 82041044). C.-F.Q. was supported by the National Science Fund for Distinguished Young Scholars (81925025), the Innovative Research Group (81621005) from the NSFC, and the Innovation Fund for Medical Sciences (2019-I2M-5-049) from the Chinese Academy of Medical Sciences.
-
The authors declare no competing interests.
-
All procedures involving infectious virus were conducted in Biosafety Level 3 laboratory (BSL-3) in Beijing Institute of Microbiology and Epidemiology and approved by the Animal Experiment Committee of Laboratory Animal Center, Beijing Institute of Microbiology and Epidemiology (approval number: IACUC-DWZX-2020–002). All institutional and national guidelines for the care and use of laboratory animals were followed.














 DownLoad:
DownLoad: