-
Dear Editor,
Acute gastroenteritis (AGE) is a leading infectious cause of morbidity worldwide, particularly among children in developing countries (Mortality and Causes of Death 2016). With the introduction of rotavirus vaccines, noroviruses have been identified as a leading cause of AGE outbreaks and sporadic disease worldwide (Hall et al. 2013). The norovirus genome contains three open reading frames (ORFs). ORF1 encodes the nonstructural viral proteins including the RNA-dependent RNA polymerase. ORF2 and ORF3 encode the respective major (VP1) and minor (VP2) structural proteins. Based on phylogenetic clustering of the complete VP1 amino acid sequence, norovirus can be divided into 10 genogroups (G) (Chhabra et al. 2019) of which GI, GII, and GIV have been found among human beings. Genogroups can be subdivided into 9 GI, 27 GII, and 2 GIV genotypes (Chhabra et al. 2019). A dual-nomenclature system has been proposed for GI and GII noroviruses using partial regions of the ORF1 RNA polymerase (P)-encoding region and partial region of ORF2.
Since the mid-1990s, genogroup II genotype 4 (GII.4) noroviruses have caused the majority of outbreaks and sporadic cases worldwide. Recombination which often occurs at the ORF1/ORF2 junction and antigenic drift are the mechanism of evolution for norovirus. New GII.4 variants emerged with 2-3-year periodicity up to 2012. Afterwards, the GII.4 Sydney capsid seems to persist through recombination, with a novel recombinant of GII.4 Sydney[P16] detected in 2014 in Germany and the Netherlands, and again in 2016 in Japan, China, and the Netherlands (van Beek et al. 2018). However, there were few studies about the genotype of norovirus in sporadic gastroenteritis cases in China, recently. Here, we describe the identified genotype of norovirus in sporadic AGE cases in Beijing area, China.
From November 2018 to September 2020, a prospective study about viral etiology of AGE in pediatric outpatients was conducted in Beijing Children*s Hospital. AGE was defined as > 3 events involving loose feces, vomiting, or both within a 24-h period. A total of 605 patients each one with a fecal sample were enrolled into this study. Among them, 64% were male, 36% were female. The sex ratio (male/female) was 1.78:1. The age of patients ranged from one month to 16.0 years (median, 1.0 year). About 64.1% (388/605) cases were infants less than 1 year old.
Viral RNA was extracted from 10% fecal suspension (mixing 0.1 g feces with 1.0 mL phosphate-buffered saline (pH 7.2)). Then a commercial kit of multiplex real-time PCR for norovirus (GI and GII) and rotavirus (group A) (Shuoshi, Jiangsu, China) was used in this study. The positive samples of norovirus were amplified by conventional RT-PCR with SuperScriptTM Ⅲ One-Step RT-PCR System with PlatinumTM Taq DNA Polymerase kit (Invitrogen). Dual-typing system of norovirus was used in this study, which amplifies a partial region of ORF1 and a partial region of ORF2 (van Beek et al. 2018). The previously published oligonucleotide primers for GII viruses were MON431 (TGGACIAGRGGICCYAAYCA) and G2SKR (CCRCCNGCATRHCCRTTRTACAT). The expected PCR product size is 570 bp (Cannon et al. 2017, 2019). All PCR products were subjected to Sanger sequencing and genotypes were assigned using the Norovirus Genotyping tool (http://www.rivm.nl/mpf/norovirus/typingtool). A phylogenetic tree was constructed using the maximum likelihood method implemented in MEGA software (version 5.01) with 1000 bootstrap replicates. The sequences of the noroviruses identified in this study were deposited in GenBank (GenBank accession numbers: MW205541-MW205668).
Overall, norovirus was identified in 30.7% (186/605) of all tested samples. The AGE patients were divided into three age groups: 0-1 year (n = 388), 2-5 years (n = 126) and 6-18 years (n = 91). The positive rate of norovirus was 34.3% in 0-1 year age group, followed by 27.0% and 20.9% in 2-5 years age group and 6-18 years age group, respectively. There were significant differences in the positive rate of norovirus between different age groups (P = 0.026). Young children are the risk groups of norovirus infection. As in this study, the detection rate of norovirus in young children is relatively high, especially in children under 1 year old.
The epidemic of norovirus is seasonal. Most outbreaks occurred during the winter season (November-March) (Jin et al. 2020). In this study, norovirus positive cases were also mainly in winter, accounted for 76.34% (142/186) of all positive cases, which is consistent with the seasonal distribution of norovirus.
For norovirus positive cases, most of them were GII strain with a positive rate of 95.7% (178/186). GI strain was only detected in 9.1% (17/186) cases. There were also 4.8% (9/186) cases with a co-infection of GI and GII strains.
Among the GII positive cases, 128 were further genotyped. GII.P31 (52.3%, 67/128) was the most common RdRp genotype, followed by GII.P16 (28.1%, 36/128) and GII.P12 (14.8%, 19/128) (Fig. 1A). For VP1 genotype, the three most common were GII.4 (68.8%, 88/128), GII.3 (14.8%, 19/128) and GII.2 (11.7%, 15/128) (Fig. 1B). The most prevalent strain was recombinant GII.4[P31] (52.3%, 67/128), followed by GII.4[P16] (16.4%, 21/128), GII.3[P12] (14.1%, 18/128) and GII.2[P16] (12.5%, 16/128). GII.17[P17] norovirus was only detected in 1.6% (2/128) cases. However, the rare recombinant norovirus genotype GII.6[P7] in China was found in 3.1% (4/128) cases here.
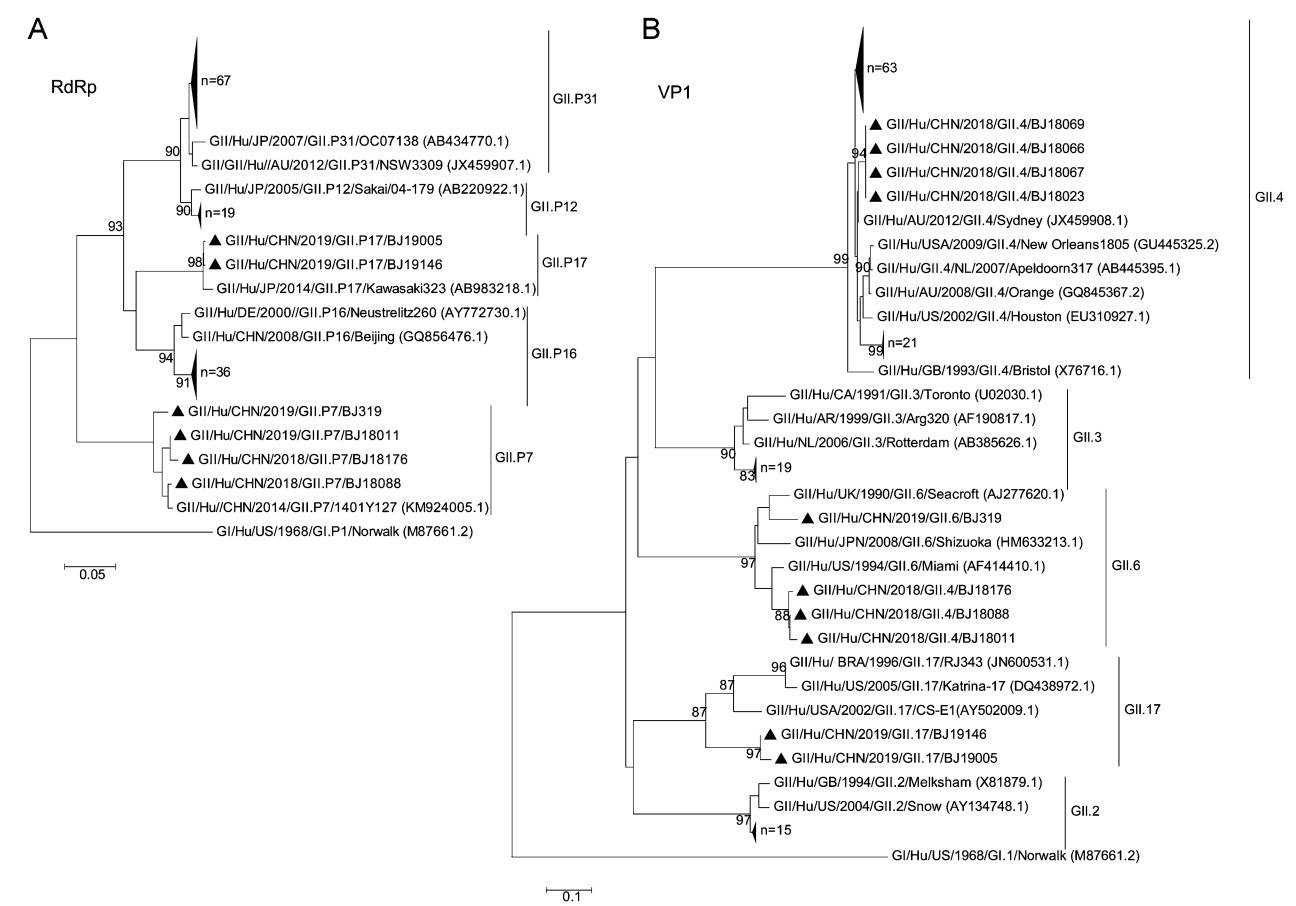
Figure 1. Phylogenetic trees of the norovirus GII partial-nucleotide sequences. A Analysis of the RNA-dependent RNA polymerase (RdRp) region (251 bp). B Analysis of the major capsid protein VP1 region (302 bp). Trees were generated by using the Neighbor-Joining method with 1,000 bootstrap replicates implemented in MEGA5 (https://www.megasoftware.net). Bootstrap values > 80 are indicated at the nodes. Strains of sufficient nucleotide sequence length were included in the trees (denoted individually with black triangles and in groups with large triangles). Reference strains are shown with accession numbers (in parentheses). Scale bars indicate nucleotide substitutions per site
Norovirus continues to evolve through genetic mutation and recombination. It is reported that, the prevalent genotype of norovirus circulating in the same region will change every several years (Hasing et al. 2019). Even in the same period, the genotypes of norovirus circulating in different regions may be different. In this study, the most prevalent norovirus was recombinant GII.4[P31] genotype in sporadic AGE children. It is similar to the study of sporadic cases in Shanghai from 2014 to 2018, in which the predominant genotype was GII.4 Sydney[P31] (Wang et al. 2019). However, it is different with norovirus outbreaks studies in the past 5 years. In China, the predominant genotype of norovirus outbreaks were GII.17[P17] and GII.2[P16] in 2014-2015 and 2016-2018, respectively (Ao et al. 2017; Gao et al. 2019; Jin et al. 2020).
In this study, the most prevalent genotype was recombinant GII.4[P31]. It is reported that over the past 10 years, GII.4 Sydney[P31] and GII.4 Sydney[P16] were the predominant strain epidemic globally (van Beek et al. 2018). GII.4 Sydney[P31] has been the predominant strain in Canada (Hasing et al. 2019) and Thailand (Chuchaona et al. 2019) since 2012. And GII.4 Sydney[P16] has been popular in Germany, Netherlands, America, Japan and Canada since 2014 (Cannon et al. 2017; van Beek et al. 2018; Hasing et al. 2019).
In conclusion, norovirus was identified in 30.7% of sporadic AGE children. And the most prevalent genotype was recombinant GII.4[P31] in Beijing area in 2018-2020. Norovirus is evolving by recombination and antigenic drift. Thus, continuous monitoring of the circulating genotypes in pediatric population is needed for current norovirus vaccine development.
HTML
-
This study was approved by the Medical Ethics Committee of the Beijing Children's Hospital, Capital Medical University, and informed consent was obtained. We would like to acknowledge all the physicians and participants for collecting clinical specimens from the patients used in this study. We thank all the reviewers for comments that improved this manuscript. This work was supported by grants from National Major Science & Technology Project for Control and Prevention of Major Infectious Diseases in China (2018ZX10201002-009-005).
-
The authors declare that they have no conflict of interests.
-
This study conformed to the 1975 Declaration of Helsinki guidelines and was approved by Medical Ethics Committee of Beijing Children's Hospital, Capital Medical University (2018-k-147).














 DownLoad:
DownLoad: