-
Dear Editor,
A novel coronavirus disease (COVID-19) caused by acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first broke out in Wuhan, Hubei Province, China, in 2019. More than 50 million cases of COVID-19 have been confirmed globally with 1,280,868 deaths reported as of November 12, 2020 (https://covid19.who.int/). RNA detection based on real-time reverse transcription polymerase chain reaction (rRT-PCR) assay has been developed and is widely used as the standard of diagnosing COVID-19 (Chinese Center for Disease Control and Prevention 2020). SARS-CoV-2 RNA can be detected from various types of specimens, with respiratory specimens mostly preferred (Pan et al. 2020; Wang et al. 2020; Zou et al. 2020). However, more data are needed to understand the profiles of viral shedding in different types of specimens from COVID-19 patients with different clinical manifestations. Antibody detection plays an important role in identifying the clinical status and outcome of a patient. Although several studies on antibody detection in COVID-19 patients have been published recently (Burbelo et al. 2020; Demey et al. 2020; Lee et al. 2020; Li et al. 2020; Liu et al. 2020; Nie et al. 2020; Okba et al. 2020; Wan et al. 2020; Xiang et al. 2020; Zhao et al. 2020; Wang et al. 2020a, 2020b, 2020c), knowledge gaps regarding the profile, dynamics, and magnitude of the antibody response in COVID-19 patients with different clinical manifestations still exist.
To understand the profiles of SARS-CoV-2 RNA and antibodies in inpatients with COVID-19, we enrolled 53 COVID-19 inpatients admitted to hospitals in Qingdao between January and March of 2020. The day of symptom onset (fever, cough, or fatigue, etc.) was defined as day 0 for most cases in the subsequent analyses. Specimens were collected in January and February on the basis of easy access by the Qingdao Municipal Center for Disease Control and Prevention according to the Technical Guidelines for COVID-19 Laboratory Testing (China CDC 2020).
The median age of the patients was 35 years old (range, 5–70 years old), and 43% of the patients were male. Of all patients, 3 (6%), 11 (21%), 33 (62%), and 5 (9%) exhibited asymptomatic, mild, moderate, and severe clinical symptoms, respectively, and clinical information was lacking for one patient. The mean duration between symptom onset and admission for all patients was 3 days (range, 0–22 days). Patients have stayed in hospital for an average of 11 days, and three patients were hospitalised for more than three weeks, with a maximum stay of 49 days. 187 specimens were collected and tested, including 142 tests for viral RNA using six types of samples (nasopharyngeal swabs, sputum, faeces, urine, blood, and conjunctival swabs).
To determine viral RNA levels in samples, real-time RT-PCR was performed using the nucleocapsid gene as a target (Lu et al. 2015; Niu et al. 2020; Wang et al. 2020). Nasopharyngeal swab and sputum samples had the highest viral loads, up to 2.9 × 106 copies/mL (mean, 1.6 × 106 copies/mL) and 1.3 × 106 copies/mL (mean, 1.1 × 106 copies/mL), respectively, substantially higher than the maximum load of 231 copies/mL (mean, 54 copies/mL) recorded in faecal specimens (Fig. 1A, left). Most nasopharyngeal swabs (95%, 18 of 19 tested) and all sputum samples (15 tested) had detectable viral loads in week 1, compared with 22% of the faecal samples (Fisher's exact test, P < 0.001; Fig. 1A). Positive test rates did not differ among the three types of samples starting in week 2 after symptom onset (Fig. 1A, left). No positive results were obtained from urine (n = 13), blood (n = 17), or conjunctival swab (n = 10) samples.
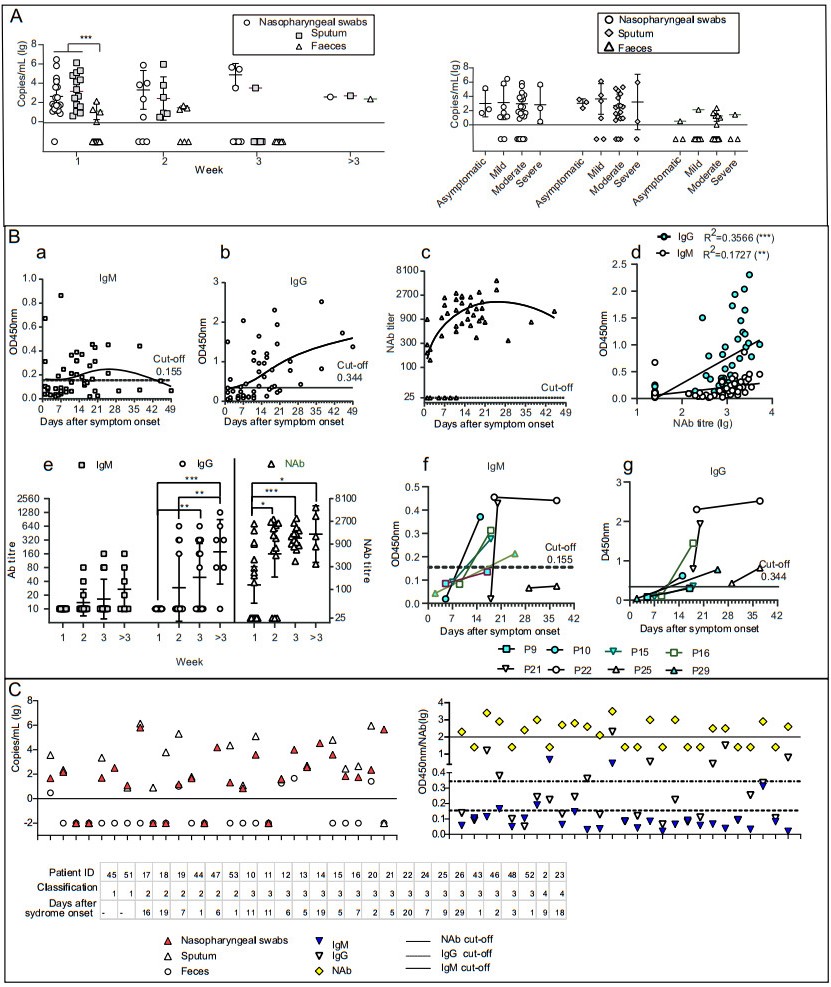
Figure 1. Detection of viral RNA and antibodies in samples of COVID-19 patients. A Comparison of viral RNA loads in COVID-19 patients detected by nasopharyngeal swab, sputum, and faecal samples (left) Viral RNA loads in COVID-19 patients with different clinical manifestations. B Serum IgM and IgG profiles in COVID-19 patients as analysed using RBD-based enzyme-linked immunosorbent assay (ELISA). a–c Cross-sectional profiles of absorbance at 450 nm (OD450nm) by IgM (a) and IgG (b) against RBD and by neutralising antibody (NAb) in a pseudovirus particle neutralisation test (c). Each dot represents an individual serum sample. d, Comparison of IgM, IgG, and NAb. e, Chronological changes (weeks after symptom onset) in IgM and IgG titres based on RBD-ELISA, and in NAb titres. f and g Changes in IgM (f) and IgG (g) levels between matched pairs of serum samples from COVID-19 patients. Each line represents an individual patient. C Detection of viral RNA and antibodies in SARS-CoV-2-infected patients. Results of matched-pair analysis to detect RNA (left) and antibody levels (right) in SARS-CoV-2-infected individuals. Classification: 1 indicated as asymptomatic patients, 2 indicated as mild patients, 3 indicated as moderate patients, 4 indicated as severe patients. *P < 0.05, **P < 0.01, ***P < 0.001.
Viral loads of nasopharyngeal swab did not differ in COVID-19 patients with different illness stage, nor that of sputum, or faecal samples. The positive rate from nasopharyngeal swab samples decreased significantly, to 43%, in week 3 after symptom onset compared to the rate in week 1 (Fisher's exact test, P < 0.05). Similarly, positive rates from sputum samples decreased significantly, from 100% to 14%, from both weeks 1 and 2 to week 3 after symptom onset (Fisher's exact test, P < 0.001 and P < 0.01, respectively). Furthermore, the average viral loads in COVID-19 patients with different clinical manifestations were assessed (Fig. 1A, right). For nasopharyngeal swab, sputum, and faecal samples, the average viral loads did not differ among patients (Fig. 1A, right). However, nasopharyngeal swabs collected from patients with mild disease status produced higher positive test rates compared with faecal samples (Fisher's exact test, P < 0.05). The proportion of positive tests differed more markedly among nasopharyngeal swab, sputum, and faecal samples collected from patients with moderate disease status (Fisher's exact test, P < 0.01).
To determine the antibody levels in serum samples, an enzyme-linked immunosorbent assay (ELISA) was performed in-house by coating ELISA plates with commercial RBD (Sino Biological Inc., Beijing, China) to detect immunoglobulin (Ig) M and IgG serum samples. The cut-off value was set at three standard deviations above the mean value of the negative cohort. A SARS-CoV-2 pseudovirus particle neutralisation test (ppNT) was performed to detect neutralizing antibody (NAb) as previously described (Nie et al. 2020; Wang et al. 2019, 2016). The median seroconversion times for IgM, IgG, and NAb were 14 (1–37), 16 (1–49), and 14 (1–45) days after symptom onset, respectively. Seroconversion of IgM, IgG, and NAb were observed in 21, 28 and 41 serum samples, respectively, that of both IgM and IgG in 18 serum samples, that of both IgM and NAb in 19 samples, and that of both IgG and NAb in 26 samples (Fig. 1B-a, -b and -c). Both IgM and IgG titres were significantly correlated with NAb (P < 0.01 and P < 0.001, respectively; Fig. 1B-d). Overall, IgM conversion exhibited an increase–decrease pattern (Fig. 1B-a), with 18% of the serum samples (n = 17) testing positive in week 1 and 54% (n = 13), 50% (n = 14) in weeks 2 and 3, respectively. After week 3, seroconversion of IgM decreased to 43% (n = 7). IgG and NAb conversion exhibited a continually increasing trend. IgG had a low conversion rate of 24% (n = 17) in week 1 and 100% (n = 14) at 3 weeks later (Fig. 1B-b). In week 1, 53% (n = 17) of the samples tested positive for NAb, increasing to 77% (n = 13) in week 2, and 100% thereafter. The average IgM titre was low throughout the observation period, whereas the average IgG titre continued to increase at 3 weeks after symptom onset (Fig. 1B-e). IgG titres increased significantly in week 3 compared to week 1 (P < 0.01), and that were significantly higher after week 3 than that in week 1 and week 2 (P < 0.001 and P < 0.01). NAb displayed special pattern with significantly higher levels in week 2 after symptom onset than that in week 1 (P < 0.05), while NAb titers in week 3 and 4 after symptom onset were not significantly higher than that in week 2 (Fig. 1B-e). IgM and IgG levels in the serum of eight patients were compared in a matched-pair analysis (Fig. 1B-f, -g). The dynamic changes of serum antibody titres exhibited diverse trends and occurred in most of the patients at 2 weeks after symptom onset. However, low IgM levels and a slow response was observed in one individual, and IgM levels in another patient decreased at 5 weeks after symptom onset, when a second serum sample was collected. By contrast, IgG levels in the second serum samples were significantly higher than in the first ones. The median seroconversion times were 14 (IgM and NAb) and 16 (IgG) days after symptom onset, similar with the results in other studies (Demey et al. 2020; Imai et al. 2020; Lee et al. 2020; Liu et al. 2020; Lou et al. 2020; Norman et al. 2020; Okba et al. 2020; To et al. 2020).
In this single centre study, we investigated the profiles of SARS-CoV-2 RNA and antibodies of 53 COVID-19 patients with different clinical manifestation (Garbati et al. 2016; Lou et al. 2020; Xiang et al. 2020). The results proved that SARS-CoV-2 excretion via the respiratory tract was the main viral source, and the profiles of SARS-CoV-2 RNA and antibodies were not related with clinical manifestation. Notably, the dynamics of SARS-CoV-2 RNA were similar in asymptomatic COVID-19 patients with others, and this aspect should be studied in further detail. Our analysis of viral loads confirmed that some asymptomatic cases can host extraordinarily high viral loads, implying that asymptomatic patients who engage in social contact can have high transmission potential (Pan et al. 2020). These cases should be managed early to prevent disease transmission (Joint Taskforce on Control 2020).
Although IgG and NAb levels increased over time, indicating seroconversion, no significant differences in IgG and NAb levels were found among COVID-19 patients with different clinical manifestations throughout the observation period. These results contradict previous studies (Long et al. 2020; Okba et al. 2020; Ozturk et al. 2020; Wang et al. 2020b, 2020c), considering the difference of longitudinal study and cross-sectional survey.
In this study, the matched-pair analysis indicated that the occurrences of the virus and antibodies were not mutually exclusive, suggesting that the development of antibodies in serum was not followed by the rapid elimination of viral RNA from the lung. NAb normally includes IgA, which is secreted in respiratory fluids and saliva (Hsueh et al. 2004; Norman et al. 2020; Okba et al. 2020). Further work by our group will focus on IgA detection.
A major limitation of this study was the limited number of patients enrolled from a single centre. Multicentre, multi-national, prospective studies on the profiles and dynamics of SARS-CoV-2 RNA and antibodies are urgently needed for prediction and diagnosis of the clinical progress and outcomes in COVID-19 patients. In addition, commercial antibody assays with the best performance in clinical settings should be chosen in future studies.
HTML
-
This work was supported by the National Key Program for Infectious Disease of China (2018ZX10101002), the National Key Research and Development Program of China (2016YFD0500301, 2020YFC0840900).
-
The authors declare no competing interests.
-
All procedures performed in studies involving human participants were in accordance with the policy on public health investigations of emerging infectious diseases issued by the National Health Commission of China and its later amendments or comparable ethical standards.














 DownLoad:
DownLoad: