-
Introduction Baculoviruses are insect-specific large double-stranded DNA viruses, which have a wide range of applications in biological control, foreign genes expression and gene delivery. As eukaryotic expression vectors, baculoviruses have advantages in expression of high level of proteins, insertion of large size foreign DNAs, as well as posttranscriptional modifications (Kost and Kemp 2016; Chambers et al. 2018). A huge number of exogenous proteins have been successfully expressed by baculovirus/ insect expression system since it was first established in 1983 (Smith et al. 1983). In addition, baculovirus-mediated gene transduction of mammalian cells is emerging as an avenue for gene therapy and drug discovery (Thimiri Govinda Raj et al. 2020).
During hundreds of million-year co-evolution with insect hosts (Théxzé x et al. 2011), most baculoviruses have developed a unique biphasic life cycle, which is characterized by the production of two morphologically distinct virion phenotypes, budded viruses (BVs) and occlusionderived viruses (ODVs). ODVs are embedded in occlusion bodies (OBs), which provide stability and environmental protection to ODVs. After being ingested by insect hosts, ODVs are released from OBs due to the alkaline environment in insect midgut and an endogenous alkaline protease, leading to a primary infection of epithelial cells. Subsequently, progeny BVs are produced, bud from infected cells and disseminate infection to permissive cells and tissues resulting in a systemic infection and death of the infected hosts (Blissard and Theilmann 2018).
When baculoviruses are used for biological control of pests, the infectivity of both ODVs and BVs is required for the primary and the secondary infection, respectively. In contrast, when baculoviruses are used as expression or gene delivery vectors in vitro, only the infectivity of BVs is required. To date, total genome sequencing of more than 172 baculoviruses has been accomplished (Wennmann et al. 2018), revealing a genome size range between 80 and 180 kb and encoding approximately 90–180 genes. Studies on comparative genomics revealed 38 core genes conserved in all of the baculoviruses (Garavaglia et al. 2012; Javed et al. 2017). These core genes are indispensable for the complete life cycle of baculoviruses. Among these core genes, some are not required for BV production, such as the per os infectivity factors (PIFs), which are only essential for oral infection mediated by ODVs (Wang et al. 2017). So far, 10 PIFs have been identified, and deletion of any had no impact on BV productions (Wang et al. 2019). As for the whole baculovirus genome, there are likely more genes which are not required for infectious BV production. Removing genes dispensable for BV production could generate a smaller baculovirus genome that theoretically should be a better vector for the insertion and expression of exogenous genes.
Synthetic biotechnology provides a powerful tool for genome wide functional studies, and the success of synthesizing a minimal bacterial genome was a hall mark (Hutchison et al. 2016). Recently, a synthetic baculovirus was made in our lab, which exhibited similar biological properties as its parental Autographa californica nucleopolyhedrovirus (AcMNPV), the type species of baculovirus (Shang et al. 2017). AcMNPV, the best-studied baculovirus, encodes 155 open reading frames (ORFs) and the functions of most genes have been successively uncovered. Review of the literatures identified 42 ORFs in AcMNPV had not been functionally verified by geneknockout assay before we initiated this study. Therefore, we systematically analyzed the impacts of these ORFs on infectious BV production by constructing gene-knockout bacmids and subsequently conducting transfection and infection assays. In addition, we determined the one-step growth curves of the knock-out viruses, aiming for future construction of an AcMNPV vector with a reduced genome size while retaining the properties of proper BV production.
HTML
-
Spodoptera frugiperda 9 (Sf9) cells were propagated at 27 C in Grace's insect medium (pH6.0; Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL). The AcMNPV bacmid bAcMNPV-egfp, which contains an egfp gene was generated in our laboratory (Zhang et al. 2020) and the corresponding virus AcMNPV-egfp was used as the control in this study.
-
To generate knockout bacmids, target genes were replaced with a Zeocin resistance gene through λ-red homologous recombination in Escherichia coli, as described previously (Hou et al. 2002). About 450-bp upstream and downstream fragments were amplified by PCR from bAcMNPV-egfp, and a 1215-bp Zeocin cassette was amplified from pIZ/V5- his vector (Invitrogen). Then, approximately 2100-bp linear fragments were acquired by overlap PCR containing upstream and downstream sequences flanking each side of Zeocin cassette, which were later used to transform L-arabinose-induced E. coli TOP10 competent cells containing bAcMNPV-egfp and the k-Red recombinase-encoding plasmid pCas9 (Jiang et al. 2015). Positive clones were selected with Zeocin/kanamycin and further verified by PCR. The resulting knockout bacmids were named as bAc△X where X represents the ORF number of the target gene.
-
To construct the repaired recombinant bacmids, the Bac-toBac method was used (Invitrogen). For those genes which had impact on infectious BV production when deleted, each ORF was PCR amplified containing its native promoter and cloned into the pFastBacDual transfer vector. The donor plasmids were individually transformed into DH10B competent cells containing the corresponding knockout bacmids and a helper plasmid expressing transposase. Recombinant bacmids were selected by gentamicin, tetracyclin and kanamycin resistance and bluewhite screening, and further identified by PCR.
-
To generate knockout and repaired recombinant viruses, five micrograms of each bacmid DNA was used for transfecting Sf9 cells with 10 lL of Cellfectin II according to the instruction manual (Invitrogen). Supernatants were harvested at 96 h post transfection (p.t.) and then inoculated to a fresh batch of healthy Sf9 cells. Transfections or infections were monitored by fluorescence microscopy. For the knockout viruses that produced infectious progeny virions and the repaired viruses, BV titer (median tissue culture infective dose, TCID50) was determined by endpoint dilution assay in Sf9 cells.
-
Sf9 cells (3 × 106) were infected with the control virus AcMNPV-egfp, knockout and repaired recombinant viruses at a multiplicity of infection (MOI) of 5 TCID50 units unless otherwise specified. Cells were incubated with viruses at 27 ℃ for 1 h, then washed three times with 2 mL of Grace's medium after inoculum was removed.
Another 2 mL of fresh Grace's medium (supplemented with 10% fetal bovine serum) were added and incubated further. Supernatants were collected at 0, 24, 48, 72, and 96 h post infection (p.i.). The BV titers were determined by an endpoint dilution assay done in triplicates and the onestep growth curves were made using Graphpad Prism software.
-
The comparison between the growth curve of WT virus and that of the recombinant virus was statistically analyzed by one-way analysis of variance (one-way ANOVA) using BV titres through 0–96 h p.i. The P value was used to represent the statistical difference. P > 0.05, the difference was not significant; 0.01 < P < 0.05, significant; P < 0.01, highly significant.
Cells and Bacmids
Construction of Knockout Bacmids
Construction of Repaired Recombinant Bacmids
Transfection and Infection Assays
One-step Growth Curves
Statistical Analysis
-
AcMNPV encodes 155 ORFs, the functions of most have already been elucidated but prior to our study, 42 ORFs were still functionally unverified by gene-knockout assays (Table 1). To cast light on their roles in BV production, we constructed gene-knockout bacmids (Fig. 1A).
Gene Knockout bacmid BV production (P value*) Category ac12 bAc△12 P > 0.05 Dispensable ac19 bAc△19 P > 0.05 Dispensable ac26 bAc△26 P > 0.05 Dispensable ac29 bAc△29 P > 0.05 Dispensable ac33 bAc△33 P > 0.05 Dispensable ac42 bAc△42 P > 0.05 Dispensable ac44# bAc△44-45 P > 0.05 Dispensable ac45# bAc4△4-45 P > 0.05 Dispensable ac47 bAc△47 P > 0.05 Dispensable ac48 bAc△48 P > 0.05 Dispensable ac52 bAc△52 P > 0.05 Dispensable ac55 bAc△55 P > 0.05 Dispensable ac56 bAc△56 P > 0.05 Dispensable ac57 bAc△57 P > 0.05 Dispensable ac58# bAc△58-59 P > 0.05 Dispensable ac59# bAc△58-59 P > 0.05 Dispensable ac60 bAc△60 P > 0.05 Dispensable ac63 bAc△63 P > 0.05 Dispensable ac72 bAc△72 P > 0.05 Dispensable ac74 bAc△74 P > 0.05 Dispensable ac84 bAc△84 P > 0.05 Dispensable ac85 bAc△85 P > 0.05 Dispensable ac87 bAc△87 P > 0.05 Dispensable ac91 bAc△91 P > 0.05 Dispensable ac97 bAc△97 P > 0.05 Dispensable ac111 bAc△111 P > 0.05 Dispensable ac112# bAc△112-113 P > 0.05 Dispensable ac113# bAc△112-113 P > 0.05 Dispensable ac116# bAc△116-117 P > 0.05 Dispensable ac117# bAc△116-117 P > 0.05 Dispensable ac118 bAc△118 P > 0.05 Dispensable ac121# bAc△120-122-REP120 P > 0.05 Dispensable ac122# bAc△120-122-REP120 P > 0.05 Dispensable ac140 bAc△140 P > 0.05 Dispensable ac149 bAc△149 P > 0.05 Dispensable ac154 bAc△154 P > 0.05 Dispensable ac13 bAc△13 0.01 < P < 0.05 Important ac51 bAc△51 P < 0.01 Important ac120# bAc△120-122 0.01 < P < 0.05 Important ac62 bAc△62 NA Essential ac82 bAc△82 NA Essential ac106/107 bAc△106/107 NA Essential *P value was the statistical analysis result of the one-step growth curve of the knockout virus in comparison with that of the control virus AcMNPV-egfp and the statistical analyses were analyzed by one-way analysis of variance (one-way ANOVA) using BV titers through 0 h to 96 h p.i.
#Those genes were analyzed using a jointly-deleted bacmidTable 1. Summary of the impact to BV production by the knockout of the 42 genes.
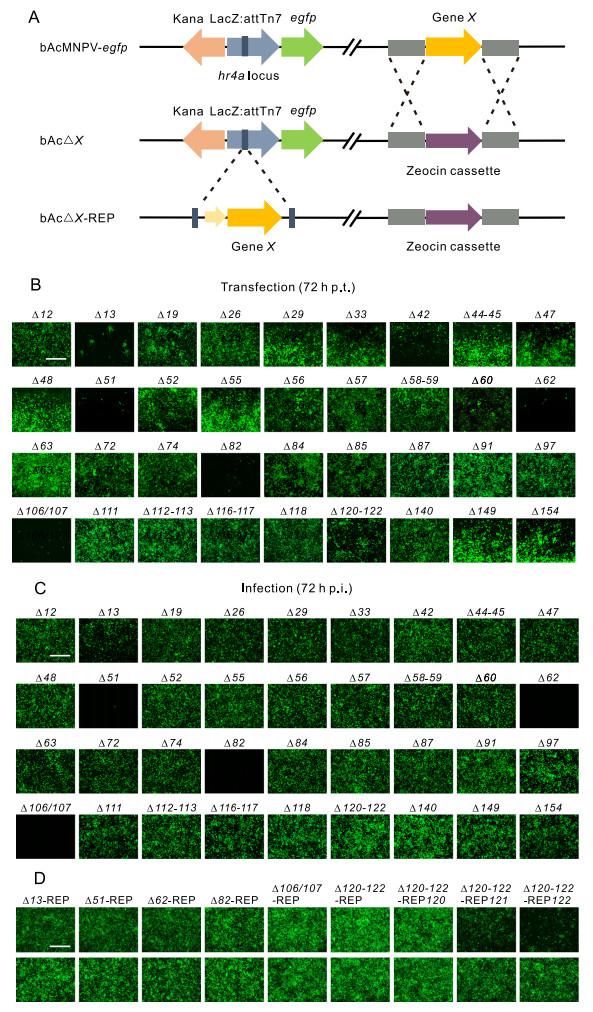
Figure 1. Construction and characterization of knockout and repaired AcMNPV bacmids. A Schematic diagram of the parental and recombinant bacmids. Knockout bacmids were constructed by replacing either the entire or partial ORF with a Zeocin cassette via homologous recombination in E. coli. For constructing repaired bacmids, each ORF driven by its native promoter was cloned into the pFastBacDual transfer vector and inserted into the attTn7 locus of the knockout bacmid by transportation. B Fluorescence microscopy of transfection of the 36 knockout bacmids. Sf9 cells were individually transfected with the knockout bacmids and the images were taken at 72 h post transfection (p.t.). Bar, 400 μm. C Fluorescence microscopy of infection of the 36 knockout bacmids. At 96 h p.t., the supernatants were used to infect Sf9 cells and the images were captured at 72 h post infection (p.i.). Bar, 400 μm. D Fluorescence microscopy of transfection and infection results of the repaired recombinants. The images were taken at 72 h post transfection (upper panel) or postinfection (lower panel). Bar, 400 μm.
As orf44 and orf45 overlap with each other, they were deleted together in the bacmid bAc△44-45; similarly, orf58 and orf59 were jointly deleted in bAc△58-59, orf112 and orf113 were jointly-deleted in bAc△112-113, orf116 and orf117 were jointly-deleted in bAc△116-117, and orf120, orf121 and orf122 were jointly-deleted in bAc△120-122. Ac106 and ac107 were initially identified as two separate ORFs but later found to be a single gene (Harrison and Bonning 2003), consequently, ac106/107 was regarded as a single gene in our study. Therefore, a total of 36 knockout bacmids were generated. All the knockout bacmids were confirmed by PCR (data not shown).
-
To determine if the deletion of the target genes were essential for BV production, the knockout bacmids and the parental bacmid bAcMNPV-egfp were analyzed by transfection and infection assays.
Fluorescence microscopy showed that at 72 h, the cells transfected with bacmids of bAc△13, bAc△51, bAc△62, bAc△82, bAc△106/107, and bAc△120-122 exhibited much less fluorescence, while the rest of the bacmids did not have an obvious effect on fluorescence expression when compared to the control bacmid (Fig. 1B). The results of subsequent infection showed that knockout of 3 genes (ac62, ac82 and ac106/107) abolished the production of infectious BVs, indicating these genes are essential for infectious BV production. For the rest of the bacmids, infectious BVs were produced, although the deletion of ac51 appeared to significantly affect the efficiency of BV production (Fig. 1C).
To confirm the impacts of the deleted genes on BV production, repaired bacmids containing ac13, ac51, ac62, ac82, ac106/107, or ac120-122 were constructed on the backbone of the relevant deletion bacmids (Material and Methods, Fig. 1A). In addition, ac120, ac121, and ac122 were individually repaired on the backbone of bAc△120- 122. Finally, 9 repaired bacmids were generated and named as bAc△13-REP, bAc△51-REP, bAc△62-REP, bAc△82- REP, bAc△106/107-REP, bAc△120-122-REP, bAc△120- 122-REP120, bAc△120-122-REP121 and bAc△120-122- REP122.
The cells transfected with bAc△13-REP, bAc△51-REP, bAc△62-REP, bAc△82-REP, bAc△106/107-REP, bAc△120-122-REP and bAc△120-122-REP120 exhibited similar fluorescence expression to that of the control bacmid at 72 h, while bAc△120-122-REP121, bAc△120-122- REP122 exhibited less fluorescence expression in comparison to that of the control virus (Fig. 1D) and subsequent infection assays showed all the repaired recombinant viruses could produce progeny viruses (Fig. 1D). The results suggest that the impact to the BV production were indeed caused by the knockout of the genes, and ac120, appears to be the key factor among ac120-122.
-
The kinetics of the knockout viruses were elucidated by one-step growth curve analyses performed with each recombinant virus except those could not produce any infectious BVs. As shown in Fig. 2A, the growth curves of vAc∆13, and vAc∆120-122 produced obviously lower yields of progeny virions from 24 h p.i. to 96 h p.i. that were significantly different from that of the control virus (0.01 < P < 0.05). In addition, vAc∆51 produced significantly lower levels of infectious BVs than those obtained with the control virus (P < 0.01), and the growth curve was performed with a low MOI of 0.2 (Fig. 2B). The rest of the knockout viruses produced levels of BV comparable to that of the WT virus (P > 0.05) (Supplementary Fig. S1, Table 1).
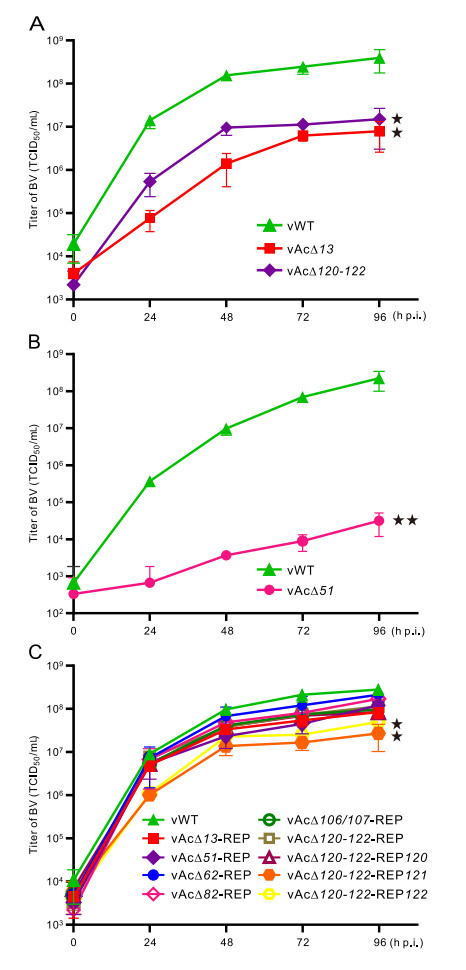
Figure 2. One-step growth curves. A One-step growth curves of vAc413, vAc4120–122 and vAcMNPV-egfp (vWT). Sf9 Cells were infected at an MOI of 5. B Growth curves of vAc451 and vAcMNPV-egfp at an MOI of 0.2. C One-step growth curves of the 9 repaired viruses and vAcMNPV-egfp. The supernatants from infected cells were collected at the indicated time points and BV titers were determined by an endpoint dilution assay in triplicates. The growth curves with significant difference to that of the vWT were indicated: ★: 0.01 < P < 0.05. ★★: P < 0.01. The results of one-step growth curves of other knockout viruses were shown in Supplementary Fig. S1, and were statistically insignificant (P > 0.05) in comparison with that of vAcMNPV-egfp (Table 1).
The kinetics of the repaired viruses were also conducted and the growth curves of vAc∆13-REP, vAc∆51-REP, vAc∆62-REP, vAc∆82-REP, vAc∆106/107-REP, vAc∆120-122-REP and vAc∆120-122-REP120 exhibited no significant difference from that of the control virus (P[ 0.05) (Fig. 2C). However, vAc∆120-122-REP121 and vAc∆120-122-REP122 produced significantly lower levels of progeny BVs than that of the control virus (0.01 < P < 0.05) (Fig. 2C).
Based on the results of transfection and one-step growth curves, we classified the target genes into three categories: (1) Dispensable: the BV production of the knockout virus is not significantly different from that of the control virus (P > 0.05); (2) Essential: the knockout bacmid was totally impaired in infectious BV production; and (3) Important: the BV production of the knockout virus is significantly different from that of the control virus (P < 0.05). According to the delineated criteria, among the 42 genes investigated, 3 genes, ac62, ac82 and ac106/107 are essential; 3 genes, ac13, ac51, ac120 are important, while the rests are dispensable for infectious BV production (Table 1, Fig. 2).
-
Following the above definition, we classify all the AcMNPV ORFs based on their impacts on infectious BV production by combining our results to previous publications. Among the 155 AcMNPV ORFs, 99 are dispensable, 46 are essential, and 10 are important for BV production. The AcMNPV genes, which are essential or important for infectious BV production are listed in Table 2, and they appear to play important roles in viral replication, transcription, structure, or regulation of host activities.
Gene Protein Function Impact on BV production (references) ac1 Proteins tyrosine phosphatase (ptp) Regulation of host activities Important Takagi et al. (1998) ac6* Lef-2 Replication Essential Wu et al. (2010) ac9 pp78/83 Structure Essential Vialard and Richardson (1993) ac10 Protein kinase-1 (PK-1) Other Essential Liang et al. (2013) ac11 Ac11 Other Essential Tao et al. (2015) ac13 Ac13 Other Important This study ac14* Lef-1, DNA primase Replication Essential Mikhailov and Rohrmann, (2002) ac24 Protein kinase interacting protein (PKIP) Nuleocapsid assembly Essential Fan et al. (1998) ac25 Single-stranded DNA binding protein (DBP) Replication Essential Vanarsdall et al. (2007a) ac34 Ac34 Other Important Cai et al. (2012) ac37 Lef-11 Replication Essential Lin and Blissard (2002) ac40* P47, RNA polymerase subunit Transcription Essential Carstens et al. (1993) ac50* Lef-8, RNA polymerase subunit Transcription Important Gauthier et al. (2012) ac51 DnaJ domain protein Other Important This study; Qiu et al. (2019) ac53* Ac53 Nucleocapsid assembly Essential Liu et al. (2008) ac53a Lef-10 Replication Essential Xu et al. (2016) ac54* VP1054 Nucleocapsid assembly Essential Marek et al. (2013) ac62* Lef-9, RNA polymerase subunit Transcription Essential This study ac65* DNA polymerase Replication Essential Vanarsdall et al. (2005) ac66 Ac66 Other Important Ke et al. (2008) ac67 Lef-3 Replication Important Nie et al. (2012) ac73 Ac73 Other Important Shao et al. (2019) ac75 Ac75 Other Essential Shi et al. (2018) ac76 Ac76 Other Essential Hu et al. (2010) ac77* Very late factor-1 (Vlf-1) Replication Essential Li et al. (2005) ac78* Ac78 Structure Essential Tao et al. (2013) ac80* GP41, tegument protein Other Essential Li et al. (2018) ac81* Ac81 Other Essential Dong et al. (2016) ac82 Telokin-like protein (TLP) Other Essential This study ac83* VP91, PIF8 Nuleocapsid assembly Essential Zhu et al. (2013) ac89* VP39 Structure Essential Thiem and Miller (1989) ac90* Lef-4 Transcription Essential Knebel-Mörsdorf et al. (2006) ac92 P33, sulfhydryl oxidase Other Essential Wu and Passarelli (2010) ac93* Ac93 Transcription Essential Yuan et al. (2011) ac94* ODV-E25 Structure Essential Chen et al. (2012) ac95* DNA helicase, P143 Replication Essential Gordon and Carstens (1984) ac98* 38K Nucleocapsid assembly Essential Wu et al. (2006) ac99* Lef-5 Transcription Essential Su et al. (2011) ac100* P6.9 Nucleocapsid assembly Essential Wang et al. (2010) ac101* BV/ODV-C42 Nucleocapsid assembly Essential Wang et al. (2008) ac102 P12 Replication Essential Gandhi et al. (2012) ac103* P45 Other Essential Yuan et al. (2008) ac104 VP80 Other Essential Marek et al. (2011) ac106/ 107 Ac106/107 Other Essential This study ac109* Ac109 Nuleocapsid assembly Essential Lin et al. (2009) ac120 Ac120 Other Important This study ac128 GP64 Structure, entry Essential Monsma et al. (1996) ac132 Ac132 Other Essential Yang et al. (2014); Fang et al. (2016) ac133* Alkaline nuclease (AN) Transcription Essential Okano et al. (2004) ac139 ME53 Replication Essential Xi et al. (2007) ac141 exon0 Other Essential Dai et al. (2004) ac142* Ac142 Other Essential McCarthy et al. (2008) ac143* ODV-E18 Structure Essential McCarthy and Theilmann (2008) ac144* Ac144 Other Essential Vanarsdall et al. (2007b) ac146 Ac146 Other Essential Dickison et al. (2012) ac153 PE38 Replication Important Milks et al. (2003) ac147-0 IE-0 Transcription ie0-ie1 is essential Stewart et al. (2005) ac147-1 IE-1 Transcription *Core gene. Table 2. AcMNPV genes essential or important for infectious BV production
Generation of Gene-Knockout Bacmids
Transfection and Infection Assay Identified 3 Genes Essential for BV Production
BV Production Property of Each Knockout and Repaired Recombinant Virus
Classification of all AcMNPV ORFs Based on Impact on Infectious BV Production
-
Genome sequencing of AcMNPV was originally published in 1994, which described a genome comprised of 133, 894 base pairs and encodes 154 ORFs of 150 nucleotides or greater (Ayres et al. 1994). Soon afterwards, several regions of the AcMNPV genome were re-sequenced and revealed that orf20/21, orf58/59, orf106/107, orf112/113 were actually joined in the C-6 strain (Harrison and Bonning 2003). In recent years, a transcriptome analysis of AcMNPV in Trichoplusia ni cells described that AcMNPV genome contains approximately 156 tightly spaced genes in which ORFs of ac106 and ac107 are contiguous and share the same 50 transcription start sites (TSS) while ORFs of ac20 and ac21, ac58 and ac59, ac122 and ac123 are discontinuous and have independent TSS (Chen et al. 2013).
In this study, we described that the AcMNPV genome contains 155 genes and considered ac106/107 as one gene. In order to identify which genes are required for BV production, we systematically knocked out 42 genes to ascertain their impacts on BV production. We found that among the genes, 36 are dispensable, 3 are essential, and 3 are important for infectious BV production. During our research, three of the previous functionally unknown genes (ac12, ac51 and ac154) of AcMNPV have been published, therefore, among the 42 genes only 39 were "functionally unverified genes". Our results corroborate with the recent data that deletion of ac12 or ac154 did not affect BV production (Costa Navarro et al. 2019; Bai et al. 2020), while deletion of ac51 resulted in significant decrease of BV production (Qiu et al. 2019).
Among the 3 essential genes, ac62 also known as lef-9, is a highly conserved gene with homologues in all baculoviruses and nudiviruses and is required for transient late gene expression (Lu and Miller 1994). Subsequently, it was shown to be a subunit of the baculovirus RNA polymerase (Guarino et al. 1998). Therefore, it is not surprising to find out a knockout thereof abolished progeny virus production. This result is also in agreement with that found with BmNPV (Ono et al. 2012).
Ac82 encodes a telokin-like protein (TLP), which is conserved in the genomes of all alpha- and betabaculoviruses. Our result showed that it is essential for infectious BV production, which is in agreement with a previous report that a temperature-sensitive TLP mutant in AcMNPV caused significant reduction in BV production (Gauthier et al. 2012). However, when its homologue bm68 was deleted from BmNPV, BV production seemed to remain normal (Iwanaga et al. 2002).
Ac106/107 homologues are found in alpha-, beta-, and gammabaculovirus genomes, but not in that of the deltabaculoviruses. It is an ORF encoding 243 amino acids and initially was identified as two separate ORFs in AcMNPV but was corrected later (Harrison and Bonning 2003). We showed here for the first time that it is essential for infectious BV production, likewise, in BmNPV, deletion of its homologue bm90 also resulted in a mutant unable to spread between cells (Ono et al. 2012). The exact role of ac106/107 in BV production needs to be further investigated.
Among the 3 genes that are important for AcMNPV BV production, ac13 encodes a 38.7 kDa protein, which has some structural similarity to some membrane proteins. Deletion of ac13 leads to nearly 40-fold decrease of BV titer at 72–96 h p.i. (Fig. 2A). Its homologue in BmNPV (bm5) was shown to encode a late protein not associated with BV or ODV (Zhou et al. 2010) and localizes in both the inner- and outer nuclear membranes and promotes formation of a virus-induced intranuclear structure (Nagamine et al. 2019).
Ac51 is a delayed-early gene encoding a 37.5 kDa DNA-J protein. We found that deletion of ac51 resulted in 2000-fold reduction of BV titer in Sf9 cells (Fig. 2B). Consistent with our results, ac51 was recently proven to be required for efficient nuclear egress of nucleocapsids and knockout thereof led to a substantially decrease in progeny BVs (Qiu et al. 2019).
Three genes, ac120-122, were shown to be important for BV production when deleted together. Subsequent repairing experiments showed that only ac120 but not ac121 or ac122 was important for BV production. Ac120 is conserved in the genomes of most alphabaculoviruses, potentially encoding a protein of 88 amino acids. An insertion/ deletion mutation of its homologue in BmNPV (bm98) had no apparent effect on infectivity (Ono et al. 2012). Further experiments are needed to explore the detail role of ac120 in BV production and to reveal why its homologue functions differently in BmNPV.
By considering our data and the previously published data, we generated a list of 155 AcMNPV genes delineating essential (46 genes) and important (10 genes) ones for infectious BV production (Table 2). To generate a minimal but efficient BV producing AcMNPV vector, all these genes need to be retained. With the help of synthetic biotechnology, we can design a so called "minimal" AcMNPV genome by removing all the dispensable genes and most homologous regions (HRs), which could reduce the size of the genome from the original 133 kb to about 68 kb (Fig. 3, Supplementary Table S1). In practice, the size of the "minimal" genome is likely to be expanded, as it is known that there are functional compensation genes in baculovirus genome. For example, deletion of either ie-1 or ie-0 supports BV production, but knockout of the both genes is lethal in AcMNPV (Stewart et al. 2005).

Figure 3. Schematic diagram of the proposed minimal AcMNPV genome. The original AcMNPV genome (left) is 133, 966 bp encompassing 155 ORFs. After removing all the dispensable genes and most of the HRs, the potential mini-genome (right) is 68, 295 bp containing 46 essential genes, 10 important genes and one HR. As double deletion of ie0 and ie1 are lethal, ac147 (encoding for both IE0 and IE1) is retained in the minimal genome. Ac121 and ac122 are retained to avoid the possible impact to ac120 as these genes are tightly adjacent to each other. Due to similar reasons, ac79, ac108, ac140 and ac145 are retained in the genome. The detailed information of the proposed mini-genome is listed in Supplementary Table S1.
It is surprising to find out that about 100 genes which occupy half of the AcMNPV genome are not required for infectious BV production, at least when they are deleted individually. As mentioned earlier, most baculoviruses have a complex life cycle with two different progeny phenotypes, BV and ODV. ODV is required for insect midgut infection, therefore, the genes solely responsible for ODV function (such as pifs) are not likely to be required for BV production. In addition, genes which are involved in manipulation of insect behavior may also be dispensable for BV production. Ac15 encodes an ecdysteroid UDPglucosyl transferase (EGT) that conjugated ecdysone with sugar molecules to arrest molting process and allow the virus to replicate in larvae (Oreilly and Miller 1989). Ac32 encodes for fibroblast growth factor (FGF), a secreted protein that stimulates insect cell motility and contributes to establishment of efficient systemic infections across host basal lamina lining midgut and tracheal cells (Detvisitsakun et al. 2005; Means and Passarelli 2010). Besides, some genes are associated with suppression of host immunity, such as ac27, ac71 and ac135 which encodes apoptosis suppressors IAP-1, IAP-2 and P35, respectively (Zeng et al. 2009). Ac27 and ac71 are dispensable for infectious BV production (Griffiths et al. 1999), while the deletion of ac135 exhibited cell-dependent effects showing that BV production was decreased significantly in Sf21 cells, but remained unchanged in T. ni (TN368) cells (Hershberger et al. 1992). Since most of the current knowledge of AcMNPV gene functions were generated from Sf9 or Sf21 cells, the information summarized in Table 2 may need to be modified when different cell lines are studied. Therefore, the impacts of different cell lines and host factors should also be considered for developing the mini-genome of baculovirus.
In summary, we systematically analyzed the impact of 42 genes on BV production in AcMNPV, and revealed 3 essential and 3 important genes. Our data delineated in this manuscript lay the foundation for designing the so far elusive "minimal" AcMNPV genome. With the aid of synthetic biotechnology, we are presently testing and remodeling the design of the minimal genome in our laboratory. In addition, our results showed that a large part of the baculovirus genome encodes genes that are not required for BV production. These genes likely play important roles in the replication cycle in larvae, such as manipulation of host behavior or suppression of host immune system. Only few of these genes have been functionally elucidated and further investigations are required.
-
This research was supported by the grants from the National Natural Science Foundation of China (No. 31872640) and the Key Research Program of Frontier Sciences of the Chinese Academy of Sciences (grant no. QYZDJ-SSW-SMC021), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11030400). The authors acknowledge colleagues from the Core Facility and Technical Support of the Wuhan Institute of Virology for technical assistance and we would like to thank Dr. Basil M. Arif for kindly editing the manuscript.
-
TC, MW and ZH designed the experiments. TC and XD performed the experiments. TC and HH analyzed the data. TC, MW and ZH wrote the manuscript. YS, YH, FD, HW, MW and ZH edited and commented on the manuscript.
-
The authors declare no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: