HTML
-
Histone deacetylases (HDACs) are enzymes that remove acetyl groups from the N-terminal lysine residues of histones, resulting in condensed chromatin and suppression of transcription (Thiagalingam et al. 2003). Histone deacetylase inhibitors (HDACIs) are promising anticancer drugs (Inoue et al. 2008; Zhou et al. 2008; Gupta et al. 2009; Chan et al. 2013). One HDACI, valproic acid (VPA), has also been shown to increase expression of human cytomegalovirus (HCMV) genes, especially immediate early (IE) genes (Michaelis et al. 2005). To identify the common effect of HDACIs on HCMV, we investigated various HDACIs and found that one of them, suberoylanilide hydroxamic acid (SAHA), had the opposite effect to VPA and inhibited HCMV replication. SAHA, which suppresses class Ⅰ, Ⅱ and Ⅳ HDACs, is approved by the U.S. Food and Drug Administration to treat malignant metastatic cutaneous T cell lymphomas (Yang et al. 2009). In addition to tumor-suppressor activity, SAHA has also been reported to increase or decrease replication of a number of viruses, including human immunodeficiency virus-1 (Klase et al. 2014), hepatitis B virus (Wang et al. 2013), hepatitis C virus (Sato et al. 2013) and Epstein-Barr virus (Daigle et al. 2011). The effect of SAHA on replication of HCMV has not, however, been previously described.
HCMV is a double-stranded DNA virus of the subfamily β-herpesvirus and has the largest genome of all herpesviruses (Davison et al. 2003; Murphy et al. 2003; Grey et al. 2005; Stern-Ginossar et al. 2012). Although initial infection with HCMV is often asymptomatic, the virus is one of the causes of hydrops fetalis and the main cause of childhood deafness and mental deficiency (Kotton 2010). A hallmark of herpesviruses is that, following the initial infection, they are able to establish a life-long latent infection in the host (Zhuravskaya et al. 1997; Pereira et al. 2007; Van Damme et al. 2015; O'Connor et al. 2016). Although the majority of people are believed to have been infected with HCMV, healthy individuals are not generally affected by reactivation of latent HCMV. This is, however a serious problem in immunocompromised patients, and HCMV can be life-threatening for organ transplant recipients and AIDS patients (Gandhi and Khanna 2004). Although some drugs are available to treat HCMV infection, there is a need for improved therapies.
HCMV is an enveloped virus, with a lipid envelope surrounding the capsid in infectious particles. At the start of HCMV infection, viral envelope proteins interact with cell surface proteins to induce cytophagy. Viral particles then fuse with the cell membrane and are transported into endosomes by vesicular transport and the viral capsid containing the viral genome is released into the cytoplasm (Compton and Feire 2007; Ciferri et al. 2015). After entering the host cell, HCMV tegument proteins suppress host gene expression and the capsids deliver the viral genome into the nucleus. Viral IE genes begin to be expressed before the virus becomes latent (White and Spector 2007). In the nucleus, the viral genome expresses delayed early (DE) genes to initiate viral DNA replication (Compton and Feire 2007). Replicated viral DNA can then transcribe viral mRNAs, which are exported to the cytoplasm and translated into late viral proteins by ribosomes (Penfold and Mocarski 1997). Late proteins initiate assembly of virions; viral proteins and the Golgi complex form a virion assembly compartment (vAC) to produce mature virions (Davison and Bhella 2007; Mocarski Jr 2007). In the vAC, virions are covered with a lipid envelope and then released from cytoplasm by vesicular transport or cell lysis (Britt 2007). The whole life-cycle of HCMV is completed within 48 h–72 h (Jean Beltran and Cristea 2014). In the present study, we found that SAHA reduced expression of HCMV IE genes. SAHA (5 μmol/L) suppressed viral DNA replication and reduced expression of the DE gene UL97. Protein levels of late genes pp28 and pp150 were also reduced by treatment with SAHA and the number of normal vACs was reduced.
Infection with HCMV can induce interferon (IFN) pathways; secreted IFNs bind to receptors on the cell surface and activate expression of IFN-stimulated genes (ISGs) (Haque and Williams 1998; Sakamoto et al. 2005). Although ISGs have important antiviral functions (de Veer et al. 2001), over long evolutionary periods some ISGs have been hijacked by viruses (Zhu et al. 2002; Schroer and Shenk 2008; Viswanathan et al. 2011; Seo and Cresswell 2013). We investigated the effect of SAHA on HCMV-infected cells by RNA-sequencing and found that SAHA suppressed HCMV-induced expression of ISGs.
RNA-sequencing data mining showed that fatty acidbinding protein 4 (FABP4) was significantly affected by SAHA treatment of HCMV-infected cells. FABPs are regulators of lipid metabolism (Coe et al. 1999), and lipid metabolism has been reported to be associated with the production of HCMV (Koyuncu et al. 2013; Seo and Cresswell 2013; Purdy et al. 2015). Here, we found that the expression of FABP4 was increased by SAHA but reduced by HCMV infection during the viral life-cycle. Knockdown of FABP4 rescued the inhibition of HCMV by SAHA, showing that FABP4 plays a role in the effects of SAHA on HCMV.
-
AD-GFP viruses were used for infection. AD-GFP is a strain of HCMV AD169 expressing green fluorescent protein (GFP). AD-GFP UV was ultraviolet-inactivated AD-GFP strain. Primary human foreskin fibroblasts (HFs) and 293 T cells were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS).
-
Two short hairpin RNAs (shRNAs) resistant FABP4 were designed, their targeting sequences and the targeting sequence of control shRNA (Ctr) were given as follows:
5'-GGAAAGTCAAGAGCACCATAA-3' (KO), 5'-CA ACAAGATGAAGAGCACCAA-3' (Ctr). Primers for overlapping PCR to construct shRNA-resisitant FABP4 cDNAs were designed as follows: 5'-CCGGGGAAAGTCAAGAGCACCATAACTCGAGTTATGGTGCTCTTGAC TTTCCTTTTTG-3', 5'-AATTCAAAAAGGAAAGTCAAG AGCACCATAACTCGAGTTATGGTGCTCTTGACTTTCC-3'; The plasmid vector (pLKO.dCMV.TetO) was used to load shRNA-resisitant FABP4 cDNAs to be transduced by lentiviral system.
-
Lentiviruses containing shRNA plasmids were assembled in 293 T cells. HFs were transduced with these lentiviruses with 5 lg/mL Polybrene, incubated at 37 ℃ for 5 h, then washed with phosphate buffer solution (PBS), and added fresh medium. At 24 h after transduction, cells were used for viral infection or drug treatment.
-
Protein levels were analyzed by Western blotting. Briefly, cells were separated from medium, washed with phosphate buffer solution (PBS), and resolved in the Sodium Dodecyl Sulfate (SDS)-containing sample buffer supplemented with protease inhibitor. Proteins from equal cell numbers were resolved by electrophoresis on an SDS-containing polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, hybridized with primary antibodies which were diluted in 5% milk or BSA, reacted with horseradish peroxidase (HRP)-conjugated secondary antibodies, incubated with ECL (Bio-Rad) to mark the secondary antibody with fluorescent. The strength of fluorescent showed the protein level.
-
HFs were cultured on coverslips (autoclaved) in 24 well plates, and separated from medium, then washed with PBS. Cells were fixed in 2% paraformaldehyde diluted with? PBS for 20 min. Then cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min, then blocked with 5% FCS in PBS for 20 min. Coverslips were incubated with 40 μL primary antibody diluted in block buffer in humidified chamber for 1 h. Then coverslips were washed with 500 μL block buffer for 5 min; then incubated with 50 μL secondary antibody diluted in block buffer for 1 h. The IE, UL38, pp28, pp65 and pp150 antibodies were provided by Dr. Jay Nelso, Dr. Dong Yu, Dr. Tomas Shenk, Dr. Min-Hua Luo and Dr. Dong Yu. The UL44, UL97, tubulin, GM130 and FABP4 antibodies were obtained from Virusy, Willget Biotech, Proteintech, Cell signaling and Proteintech. After that, coverslips were washed by PBS for 5 min, covered by 8 μL Prolong Gold, sealed with nail polish and then dried for 30 min in dark. Finally, samples were observed by confocal laser scanning microscope.
-
Titration of virus suspensions was performed on HFs. HFs were seeded on 96 well dishes with 80% confluence. The medium was diluted with virus 10 folds with DMEM, then the diluents were diluted 10 folds, like this, 7 different virus dilutions were made, and HFs were infected with different virus dilutions. Cells were placed in 37 ℃ incubator with 5% CO2. 14 days post infection, recorded the GFP positive wells by using a fluorescent microscope for TCID50 infectivity and IC90 calculating: lgTCID50 = L-D(S-0.5). L: Logarithm of maximum dilution; D: Difference in logarithm of dilution; S: Sum of positive pore ratios. IC90 is the drug concentration when TCID50 decreased 90% compared to Mock.
-
HFs were treated with DMSO or drugs each in two 96 plates wells for 3 days. Cell Counting Kit-8 (Beyotime Biotechnology) was used to test the cell quantities in each well. A450 were recorded for cytotoxity and CC50 calculating: CR = (C-R)/C*100%. CR: cytotoxity ratios; C: A450 of control cell; R: A450 of the remaining cells. CC50 is the drug concentration when CR = 50%.
-
Total RNA was extracted by using the TRIzol reagent (Invitrogen) and treated with the Turbo DNA-free reagent (Ambion) to remove genomic DNA contaminants. cDNA was reverse transcribed from total RNA with random hexamer primers using the a High Capacity cDNA reverse transcription kit (Takara). Cellular cDNA was quantified by quantitative real-time PCR (qRT-PCR) using the SYBR Advantage qPCR Premix (Takara) and primer pairs used for the viral gene were in Supplementary Table S1. The amount of each mRNA was normalized by using GAPDH as the internal control.
To measure the relative viral genome per cell, total DNA was extracted by Phenol: Chloroform: Isoamyl alcohol extraction = 1:1:1. DNA was quantified by realtime quantitative PCR using the SYBR Advantage qPCR Premix (Takara). The relative quantity of viral genome per cell was symbolized by the sequence in IE gene, and normalized using β-actin as the internal reference.
-
Total RNA was extracted by using the TRIzol reagent (Invitrogen). RNA sequencing was progressed by Shanghai Biotechnology Corporation. Results were analysed by bioinformational data mining.
-
Data are shown as means ± standard deviations (SD) and calculated using the two-tailed Student's t-test. A P-value of < 0.05 was thought to be statistical significance for each test.
Viruses and Cell Lines
Plasmids
shRNA Knockdown
Western Blotting
Immune Fluorescence
TCID50 Assay and IC90
Cytotoxity Test and CC50
Quantitative Real-Time PCR
RNA Sequencing
Statistical Analysis
-
Human fibroblasts (HFs) infected with recombinant HCMV expressing green fluorescent protein (AD-GFP) were used to test the effect of HDACIs on HCMV replication. Treatment with SAHA (10 μmol/L) reduced the GFP signal compared with the control (DMSO), indicating that SAHA inhibits HCMV replication in HFs (Fig. 1A). 4', 6-Diamidino-2-phenylindole (DAPI) was used as a nuclear counterstain. We then compared the effect of SAHA with that of other HDACIs. The cells were treated with different HDACIs and viral growth was determined by measuring the intensity of the GFP signal. SAHA (2 μmol/L) effectively inhibited HCMV replication whereas neither the pan HDACI VPA (1 mmol/L) nor the specific HDAC6 inhibitor tubacin (5 μmol/L) had any effect on HCMV replication. These data demonstrate that the effect of SAHA on HCMV replication is not common to all HDACIs (Fig. 1B).

Figure 1. SAHA inhibits HCMV at concentrations that show low cytotoxicity. A Fluorescence intensity assay used to test antiviral activity of SAHA. HFs infected with AD-GFP were treated with SAHA (10 μmol/L) or DMSO (control). DAPI was used to counterstain the nuclei. B Growth curves of HCMV treated with different HDACIs. HFs infected with AD-GFP (MOI = 0.1) were treated with SAHA (2 μmol/L), VPA (1 mmol/L) or tubacin (5 μmol/L). Supernatants were collected 2, 4 and 6 days post-infection and viral titers were measured by TCID50 assay. C Cytotoxicity of SAHA determined by CCK-8 assay. HFs were treated with SAHA for 72 h and then incubated with CCK-8 for 4 h. Relative cell quantities were measured by the value of A450. D SAHA dose-dependently inhibits HCMV replication. HFs were infected with AD-GFP (MOI = 0.1) and treated with SAHA (0, 0.5, 1 and 2 μmol/L). Supernatants were collected 6 days post-infection and viral titers were measured by TCID50 assay.
We also tested the cytotoxicity of SAHA in HFs and found that low concentrations were only slightly toxic (CC50 = 30 μmol/L, Fig. 1C). We then determined whether the inhibition of viral replication by SAHA is dosedependent, using different SAHA concentrations (0, 0.5, 1, 2 and 5 μmol/L). Inhibition of HCMV replication was more significant at higher concentrations: 2 μmol/L SAHA inhibited HCMV by > tenfold and 5 μmol/L SAHA inhibited HCMV by > 100-fold (IC90 = 1.2 μmol/L, Fig. 1D), showing that inhibition of HCMV replication by SAHA is dose-dependent. Since the IC90 against HCMV is much lower than the CC50, SAHA is safe at concentrations that effectively reduce HCMV replication.
-
Since a relatively low concentration of SAHA (2 μmol/L) reduced viral yield by > 90% (Fig. 1D), we wanted to further test its efficacy. We tested SAHA (2 μmol/L and 5 μmol/L) in HFs infected with HCMV at high and low multiplicity of infection (MOI). At MOI = 3, 2 μmol/L SAHA reduced viral replication by ~ 56.5-fold and 5 μmol/L SAHA reduced viral replication by ~ 4060-fold (Fig. 2A). At low MOI (MOI = 0.03), 2 μmol/L SAHA reduced viral replication by ~ 96.8-fold and 5 μmol/L SAHA reduced viral replication by ~ 3740-fold (Fig. 2B). These results show that HCMV infection can be significantly suppressed by SAHA.
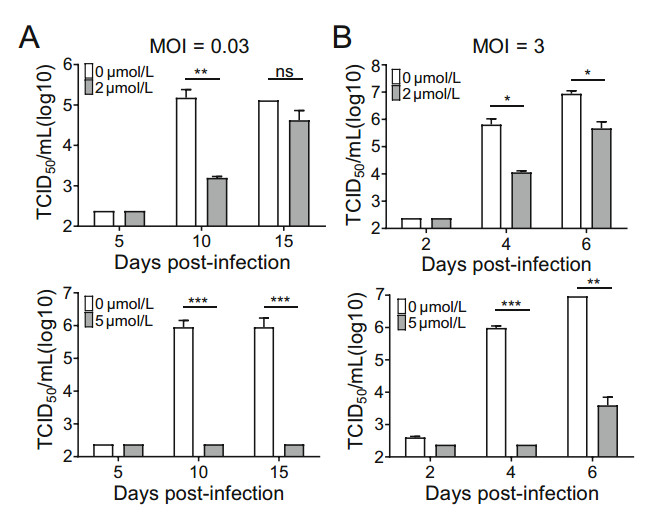
Figure 2. SAHA reduces HCMV replication. A Multi-step growth curve analysis of HCMV infection following treatment with SAHA. HFs infected with AD-GFP (MOI = 0.03) were treated with SAHA (2 or 5 μmol/L). Cell-free virus in the supernatant was collected at the indicated times after infection and viral titers were measured by TCID50 assay. B Single-step growth curve analysis of HCMV infection following treatment with SAHA. HFs infected with ADGFP (MOI = 3) were treated with SAHA (2 or 5 μmol/L). Cell-free virus in the supernatant was collected at the indicated times after infection and viral titers were measured by TCID50 assay. Statistical significance was calculated using the Student's t test (ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001).
-
To investigate which step in the HCMV life-cycle is affected by treatment with SAHA, we quantified relative viral genome number per cell and measured expression of viral genes. A lower concentration of SAHA (2 μmol/L) had little effect on viral genome replication whereas a higher concentration (5 μmol/L) reduced replication by ~ 80-fold at both 48 and 72 h post infection (hpi) (Fig. 3A). These results show that 5 μmol/L SAHA reduces the amount of HCMV genome in infected cells.
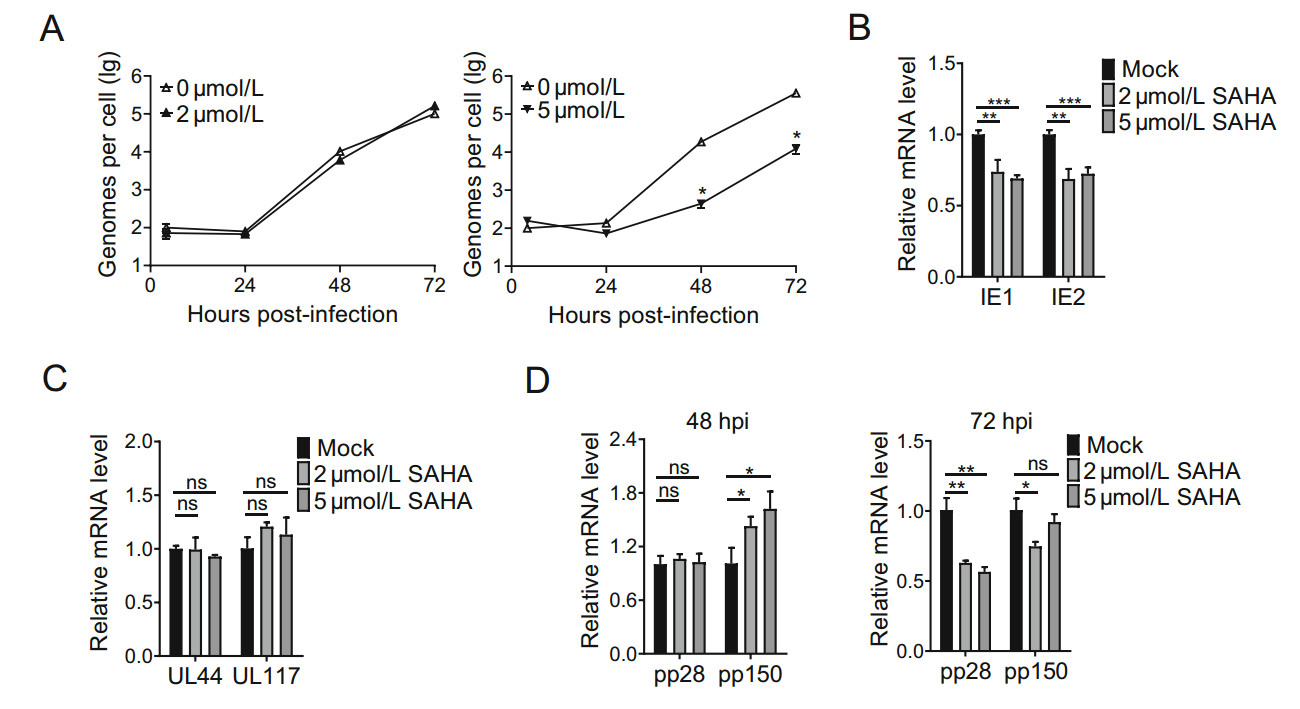
Figure 3. SAHA suppresses replication of viral DNA and transcriptions of IE genes. A qRT-PCR analysis showing relative quantities of viral genome. HFs were infected with AD-GFP (MOI = 0.3), without SAHA or with SAHA (2 or 5 μmol/L). Total DNA was collected 8 hpi and quantified by qRT-PCR of IE genes. The internal reference was β-actin. B qRT-PCR analysis of transcription levels of IE genes. HFs were infected with AD-GFP (MOI = 3), without SAHA or with SAHA (2 or 5 μmol/L). Total RNA was collected 8 h post-infection and mRNA levels of IE1 and IE2 genes were quantified by qRT-PCR. C qRT-PCR analysis of transcription levels of DE genes. Total RNA was collected 24 hpi and mRNA levels of UL44 and UL117 genes were quantified by qRT-PCR. D qRT-PCR analysis of transcription levels of late genes. Total RNA was collected 48 and 72 hpi. mRNA levels of pp28 and pp150 genes were quantified by qRT-PCR. The internal reference was GAPDH. Statistical significance was calculated using the Student's t test (ns, P > 0.05; **P < 0.01; ***P < 0.001).
The transcriptional activities of viral genes were determined by quantifying viral mRNA levels. HFs were infected with HCMV and treated with SAHA (2 or 5 μmol/L) for 8, 24, 48 and 72 h. Transcription levels of IE genes were measured at 8 hpi, and both 2 and 5 μmol/L SAHA were found to reduce transcriptions of IE1 and IE2 (Fig. 3B). This suggests that SAHA inhibits HCMV at an early point in the viral life-cycle. We measured transcriptions of viral DE genes (UL44, UL117) at 24 hpi. Unlike IE genes, UL44 and UL117 were not downregulated by treatment with SAHA (Fig. 3C). We also measured transcriptions of late genes (pp28, pp150) at 48 and 72 hpi. Transcriptions of pp28 and pp150 was not reduced at 48 hpi, whereas pp28 appeared to be reduced at 72 hpi (Fig. 3D). These results show that SAHA primarily inhibits transcriptions of HCMV IE genes.
-
The accumulation of viral proteins IE1, IE2, UL44, UL38, UL97, pp28, pp65 and pp150 was examined by immunoblotting. Both 2 μmol/L and 5 μmol/L SAHA had similar effects on accumulation of these proteins. SAHA suppressed IE2 at an early time point (8 hpi), and suppressed UL97 kinase and late proteins, pp28, pp65 and pp150, at later time points (48 or 72 hpi) (Fig. 4A). These results indicate that SAHA inhibits the expression of HCMV proteins.

Figure 4. SAHA reduces levels of HCMV viral proteins. A Western blotting was used to determine the effect of SAHA on expression of HCMV proteins. HFs were infected with AD-GFP (MOI = 3), without SAHA or with SAHA (2 μmol/L or 5 μmol/L) and cell lysates were collected at the indicated times after infection. The internal reference was tubulin. (hpi: hours post-infection). B Immunofluorescence staining showing assembly compartments of HCMV virions following treatment with SAHA. HFs were infected with AD-GFP (MOI = 0.5) without SAHA or with SAHA (2 μmol/L). DAPI was used to stain the nuclei, GM130 was used to stain the Golgi complex and pp28 was used to stain the virion assembly compartment. And the virion assembly compartment was pointed out by white arrows. Ratios of regular and irregular vACs were calculated and recorded. Statistical significance was calculated using the Student's T Test (***P < 0.001). (ND: no drug).
In infected cells, HCMV forms vACs, which recruit viral proteins and the Golgi complex to form a particle within the host nucleus, pulling the nucleus into a kidneyshape. UL97 kinase plays an important role in the formation of vACs (Azzeh et al. 2006; Sharma et al. 2015) and pp28 is a viral marker protein of vACs. Since UL97 and pp28 were suppressed by treatment with SAHA, we treated HCMV-infected cells with SAHA and observed the changes in the vACs. In the presence of SAHA, the vACs lost their regular particle shape and pull on the nucleus (Fig. 4B, left). Statistical data showed that SAHA significantly reduced the number of regular vACs per cell (Fig. 4B, right). All of these results indicate that SAHA inhibits production of HCMV viral proteins.
-
In the studies described above, SAHA effectively inhibited HCMV replication when the cells were treated with SAHA at the same time with viral infection. To determine which stage of the viral life-cycle is inhibited by SAHA, we treated cells with SAHA at different times post-infection. The HCMV life-cycle is ~ 48–72 h and we infected HFs with high MOI (MOI = 3) HCMV to ensure that all the cells were infected in a single step. We then treated the cells with SAHA every 24 h, from 48 to 120 h post-infection. SAHA reduced HCMV replication by ~ 6.6-fold when it was added 48 hpi, but had no effect when it was added 72, 96 or 120 hpi (Fig. 5). These data suggest that SAHA cannot reduce HCMV replication at late times postinfection.

Figure 5. AHA does not inhibit HCMV infection at late times postinfection. Growth curve analysis of HCMV with post-infection treatment with SAHA. HFs were infected with AD-GFP (MOI = 3), without or with SAHA (5 μmol/L) at the indicated time points postinfection for 24 h. Cell-free virus in the supernatant was collected and viral titers were measured by TCID50 assay.
-
To investigate the mechanisms underlying the antiviral effect of SAHA, we used bioinformatics to focus on relative pathways or cellular factors. We harvested HFs infected with AD-GFP and treated the cells with different concentrations of SAHA for 48 hpi, and carried out RNA sequencing. We identified many specific pathways that were significantly altered in SAHA-treated cells, one of which is associated with IFN-mediated immunity (Fig. 6A). Data mining showed that SAHA reduced many types of IFN-stimulated genes in HCMV-infected HFs (Fig. 6B).
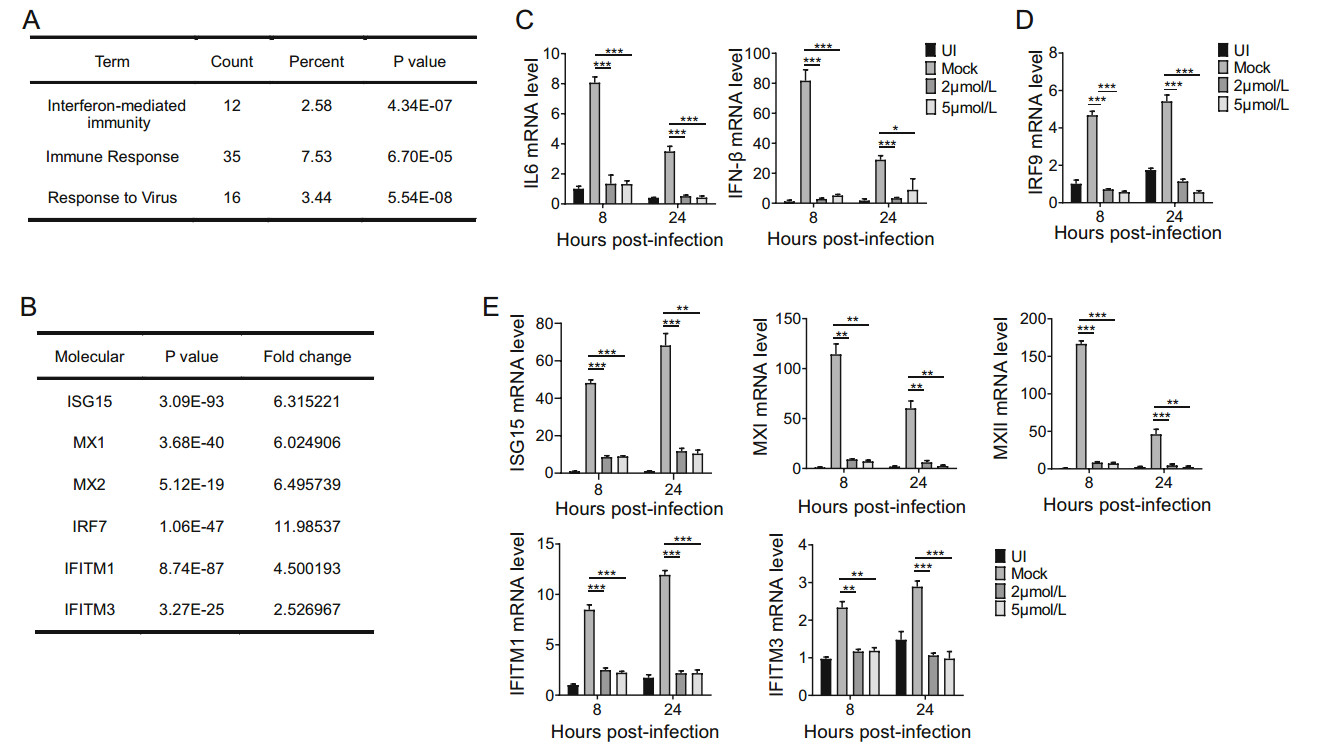
Figure 6. SAHA suppresses HCMV-induced immune response. A RNA sequencing analysis focused on pathways altered by treatment with SAHA. HFs were infected with HCMV (MOI = 3) without SAHA or with SAHA (5 μmol/L). Total RNA was collected 48 hpi. B RNA sequencing analysis focused on molecules in IFN-mediated immune pathways altered by treatment with SAHA. C Transcriptions of IL6 and IFN-β genes, measured by qRT-PCR, following treatment with SAHA. HFs were uninfected (UI), infected with AD-GFP (MOI = 3) or mock-infected and treated with SAHA (2 μmol/L or 5 μmol/L). Total RNA was collected at the indicated time points post-infection and levels of mRNA of viral genes were quantified by qRT-PCR. The internal reference was GAPDH. D Transcription of IRF9 gene, measured by qRT-PCR. E Transcriptions of ISG15, MXI, MXII, IRF7, IFITM1 and IFITM3 genes, measured by qRT-PCR. Statistical significance was calculated using the Student's t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Viral infections often induce host response to the virus; IFN-mediated immunity and the inflammatory response are the main types of innate immunity induced by viruses. We measured transcription levels of the inflammatory cytokine IL6 and IFN-β, and found that HCMV promoted transcriptions of IL6 and IFN-β genes, whereas SAHA (2 μmol/L and 5 μmol/L) significantly inhibited transcription levels (Fig. 6C). These data indicate that SAHA reduces the host innate immune response to HCMV infection.
Type Ⅰ IFNs can bind to IFN receptors to activate STAT1-, STAT2- and IRF9-mediated transcriptions of ISGs. SAHA suppressed HCMV-induced increase in the transcription of IRF9 (Fig. 6D). We also measured transcriptions of ISGs and found that, transcription levels of ISG15, MXI, MXII, IRF7, IFITM1 and IFITM3 in HCMVinfected cells were all reduced by 2 and 5 μmol/L SAHA (Fig. 6E). These data show that SAHA reduces ISGs induced by HCMV infection. Overall, we found that SAHA suppressed the HCMV-induced immune response.
-
Fatty acid metabolism has been reported to be important for HCMV infection (Koyuncu et al. 2013; Seo and Cresswell 2013; Purdy et al. 2015). FABPs, which can bind to and transport fatty acids, play important roles in fatty acid metabolism. In RNA sequencing results, SAHA influenced the transcription of FABP4 (data not shown). Here, we confirmed the results by qRT-PCR (Fig. 7A) and Western blotting (Fig. 7B) that SAHA increased the transcription and expression of FABP4, while HCMV decreased them. To investigate whether SAHA suppresses HCMV by increasing FABP4, we constructed an FABP4 shRNA to knockdown its expression. The shRNA decreased FABP4 mRNA levels (Fig. 7C) and protein levels (Fig. 7D) in HFs. FABP4-knockdown HFs were then infected with HCMV and treated with SAHA. SAHA effectively inhibited HCMV replication, and knockdown of FABP4 significantly reversed the inhibitory effect of SAHA on HCMV replication (Fig. 7E). These results show that FABP4 contributes to the effects of SAHA on HCMV replication.
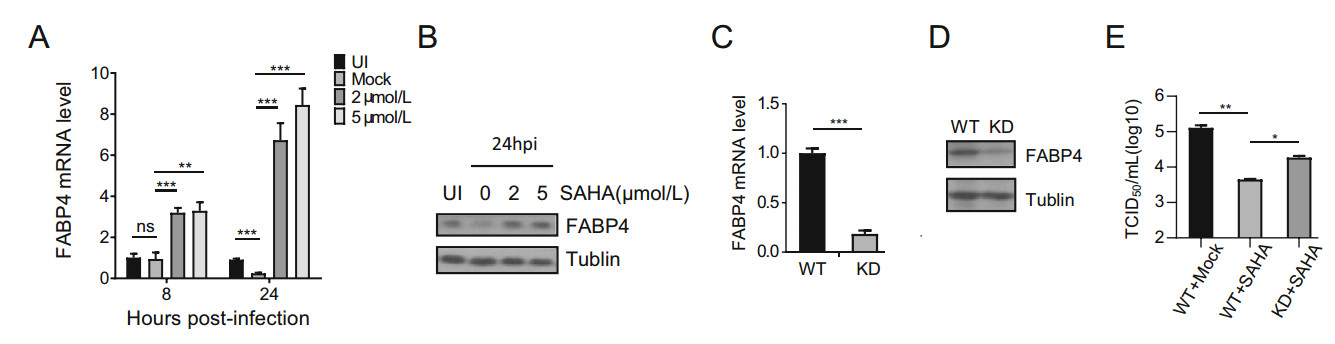
Figure 7. FABP4 contributes to inhibition of HCMV replication by SAHA. A qRT-PCR showed that SAHA increased transcriptional activity of FABP4. HFs were uninfected with AD-GFP (IU) or infected with AD-GFP (MOI = 3) without SAHA or with SAHA (2 μmol/L or 5 μmol/L). Total RNA was collected 8 and 24 h postinfection. B Western blotting showing that FABP4 protein levels were affected by SAHA. HFs were uninfected with AD-GFP (IU) or infected with AD-GFP (MOI = 3) without SAHA or with SAHA (2 μmol/L or 5 μmol/L).Cell lysates were collected 24 h postinfection. C FABP4 expression in FABP4 knock-down HFs, measured by qRT-PCR. Wild type or FABP4 knock-down HFs (2 × 106) were collected, and levels of FABP4 mRNA were quantified by qRT-PCR. D Western blotting showing FABP4 expression in FABP4 knock-down HFs. Wild type or FABP4 knock-down HFs (1 × 106) were collected, and levels of FABP4 protein were quantified by western blotting. E Growth curve analysis of HCMV infection in FABP4 knock-down cells following treatment with SAHA. Cell-free virus in the supernatant was collected at the indicated time points after infection and viral titer was determined by TCID50 assay. Statistical significance was calculated using the Student's t test (*P < 0.05; **P < 0.01; ***P < 0.001).
SAHA Inhibits HCMV Replication at Concentrations that Show Low Cytotoxicity
SAHA is Highly Effective against HCMV Replication
SAHA (5 μmol/L) Suppresses Replication of HCMV DNA and Transcriptions of IE Genes
SAHA Reduces Levels of HCMV Viral Proteins
SAHA does not Inhibit HCMV Infection at Late Times Post-Infection
SAHA Suppresses HCMV-Induced Immune Response
Fatty Acid-Binding Protein 4 Contributes to Inhibition of HCMV by SAHA
-
HDACIs are primarily used as anticancer drugs but some HDACIs have been shown to increase HCMV replication (Michaelis et al. 2004). Since HCMV poses a particular threat to immunocompromised patients and can be lifethreatening, some HDACIs may not be suitable for cancer patients infected with this ubiquitous virus. The FDA approved anticancer drug SAHA can, however, be considered in such circumstances since SAHA (2 and 5 μmol/L) significantly reduced HCMV replication in HFs, as demonstrated by median tissue culture infectious dose (TCID50) assays (Fig. 2A and 2B).
As the concentration of SAHA was increased from 2 μmol/L to 5 μmol/L, HCMV titers decreased ~ 39-fold at MOI = 0.03 and ~ 72-fold at MOI = 3 (Fig. 2A and 2B). A higher concentration of SAHA (5 μmol/L) inhibited HCMV DNA replication but a lower concentration of SAHA (2 μmol/L) did not (Fig. 3A). The mechanisms by which higher concentrations of SAHA inhibit HCMV replication require further investigation.
Licensed antiviral drugs, including ganciclovir, cidofovir and foscarnet, all affect replication of viral DNA, and thus expression of viral genes, by targeting viral pUL54 (DNA polymerase) (Biron et al. 1986; Sullivan et al. 1992; Chou 1999; Lurain and Chou 2010). We found that SAHA reduced the expression of many viral proteins, including IE, DE and late proteins (Fig. 4A), and FABP4 contributed to the effects of SAHA on HCMV replication (Fig. 7). HCMV usually remains latent in healthy individuals, whose immune systems keep the virus in check. In cancer patients, whose immune response is compromised, SAHA can be used both as an anticancer agent and to suppress reactivation HCMV.
RNA sequencing showed that many ISGs were repressed by treatment with SAHA (Fig. 6B). Over long-time evolution, some of them have been utilized by the viruses (Zhu et al. 2002; Schroer and Shenk 2008; Viswanathan et al. 2011; Seo and Cresswell 2013). We previously found that IFITMs facilitate morphogenesis of HCMV vACs (Xie et al. 2015) and here we observed that SAHA suppressed the formation of vACs (Fig. 4B). In our opinion, SAHA inhibtited HCMV replication by inducing expression of fatty acid-binding protein 4 at early phase; Meanwile, the reduction of HCMV-utilized ISGs (eg, IFITMs) caused by SAHA treatment, disrupted the normal viral life cycle, aggregated the effects of SAHA on HCMV replication.
FABPs are regulators of lipid metabolism (Coe et al. 1999). Lipid metabolism affects not only the structure of the cell membrane but also other membrane structures, including the endoplasmic reticulum membrane, Golgi membrane, lysosome membrane and nuclear membrane (Budin and Devaraj 2012; Bustin 2015). HCMV is an enveloped virus and the cellular membrane system participates in virion entry, assembly and release. The stability of the membrane system is thus clearly important for the maturation and release of virions. HCMV induces lipogenesis and it has been reported that lipid metabolism affects the production of HCMV and other enveloped viruses (Munger et al. 2008; Purdy et al. 2015). We found that SAHA increased levels of FABP4, leading to reduced viral reproduction (Fig. 7A), which may be associated with an effect on cellular membrane systems, most likely by disturbing entry of virions, exportation of viral mRNAs, and assembly or release of virions.
Many membrane components, including proteins, lipids and even sugars, contribute to the regulation of membrane function. In purely lipid terms, directly associated lipid metabolism factors, such as ASL6, ELVOL7 and FABPs (Koyuncu et al. 2013; Harjes et al. 2014; Purdy et al. 2015), and indirectly associated signaling factors all play roles in maintaining and refreshing the structure and function of membranes. Knockdown of FABP4 gene expression, which was upregulated by SAHA, only partially rescued inhibition of HCMV replication by SAHA (Fig. 7E). Possible reasons for the incomplete rescue are that SAHA likely affects other factors associated with the membrane function and that the effects of SAHA are not restricted to the cellular membrane system.
-
We thank Dr. Jay Nelso, Dr. Dong Yu, Dr. Tomas Shenk, Dr. Min-hua Luo for providing the antibodies. We thank Dr. Pei Hao for bioinformatic data mining. This research was supported by National Key R&D Program of China Grant (2016YFA0502101), National Natural Science Foundation of China (grants 81371826 and 81572002 to Z.Q., grants 31300148 and 31570169).
-
ZQ, ZL and BX designed the experiments. ZL and BX performed the experiments. ZL and ST analyzed the data. ST edited pictures. ZL and ST wrote the paper. ZQ finalized the manuscript. All authors read and approved the final version of the manuscript.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: