2021 Vol.36(6)
Human endogenous retroviruses (HERVs) are remnants of ancient exogenous retroviruses (XRV) infected in the germ line cells of our ancestors over millions of years. Nowadays, HERV has become an important part of human genome and would provide important biological functions. In this issue, Wang et al. analyzed large-scale transcriptome sequencing data of human cells infected with various viruses, including influenza A and B virus, Zika virus, hepatitis C virus, measles virus, West Nile virus, Crimean-Congo hemorrhagic fever virus, respiratory syncytial virus, rhinovirus, Epstein Barr virus, and SARS-CoV-2. They found that a variety of viral infections could induce HERV activity to varying degrees, and identified 43 shared key HERVs. Interestingly, the key HERV sites reported here are adjacent to the interferon-stimulated genes and upregulated simultaneously. These results strongly suggest that HERVs are involved in host antiviral immunity, and the HERV gene locus may be a target for antiviral drug development. The cover image describes that the antiviral power gained from the transcriptional activation of HERV loci (the shining stars) would protect humans from viral infections (kindly provided by Miao Wang and Di Liu). Please see page 1315–1326 for details.
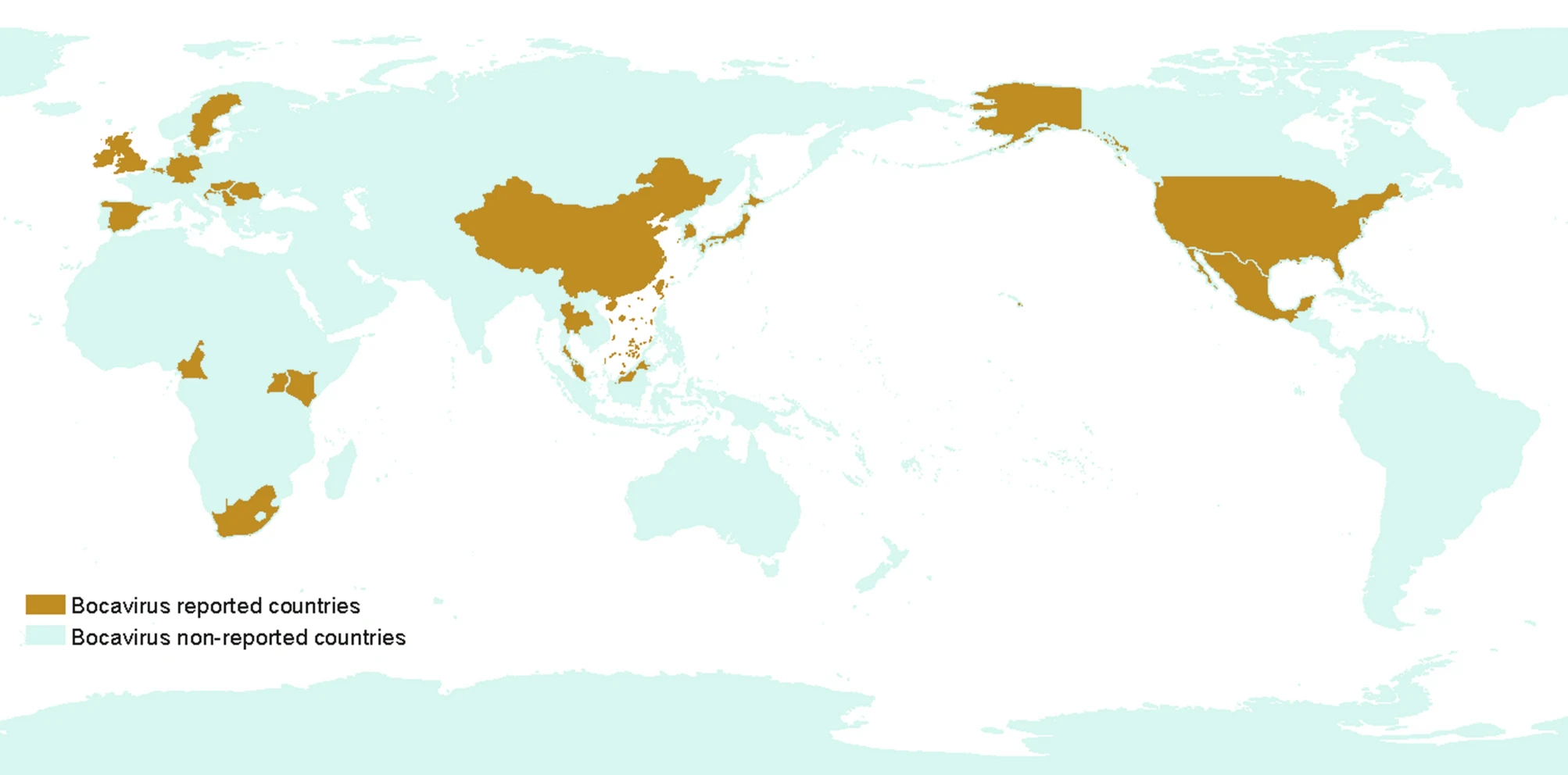
Porcine Bocavirus: A 10-Year History since Its Discovery
2021, 36(6): 1261 doi: 10.1007/s12250-021-00365-z
Porcine bocavirus (PBoV) is a single-stranded DNA virus, belongs to the genus Bocaparvovirus of family Parvoviridae. It was discovered along with porcine circovirus 2 (PCV 2) and torque tenovirus (TTV) in the lymph nodes of pigs suffering from postweaning multisystemic wasting syndrome (PMWS) in Sweden in 2009. PBoV has been reported throughout the world, mostly in weaning piglets, and has a broad range of tissue tropism. Since PBoV is prevalent in healthy as well as clinically infected pigs and is mostly associated with coinfection with other viruses, the pathogenic nature of PBoV is still unclear. Currently, there are no cell lines available for the study of PBoV, and animal model experiments have not been described. This review summarizes the current state of knowledge about PBoV, including the epidemiology, evolution analysis, detection methods, pathogenesis and public health concerns.
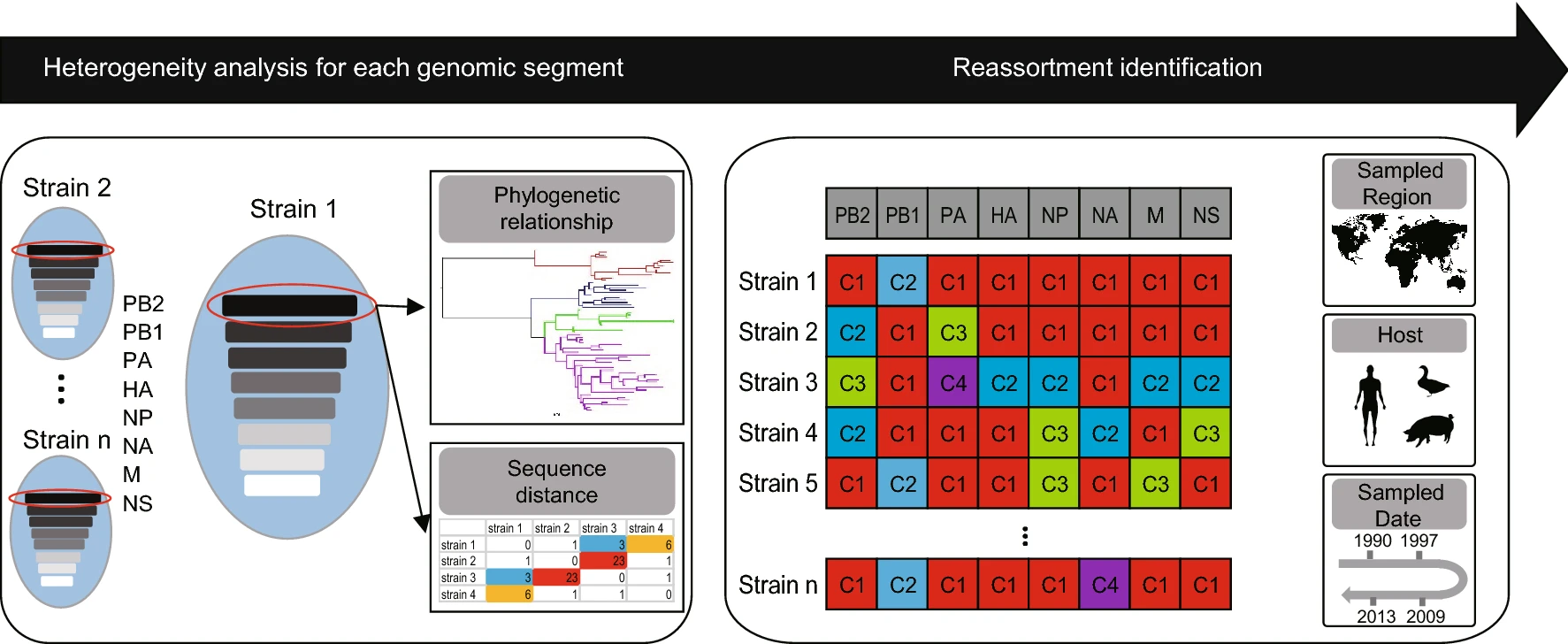
Progress and Challenge in Computational Identification of Influenza Virus Reassortment
2021, 36(6): 1273 doi: 10.1007/s12250-021-00392-w
Genomic reassortment is an important evolutionary mechanism for influenza viruses. In this process, the novel viruses acquire new characteristics by the exchange of the intact gene segments among multiple influenza virus genomes, which may cause flu endemics and epidemics within or even across hosts. Due to the safety and ethical limitations of the experimental studies on influenza virus reassortment, numerous computational researches on the influenza virus reassortment have been done with the explosion of the influenza virus genomic data. A great amount of computational methods and bioinformatics databases were developed to facilitate the identification of influenza virus reassortments. In this review, we summarized the progress and challenge of the bioinformatics research on influenza virus reassortment, which can guide the researchers to investigate the influenza virus reassortment events reasonably and provide valuable insight to develop the related computational identification tools.
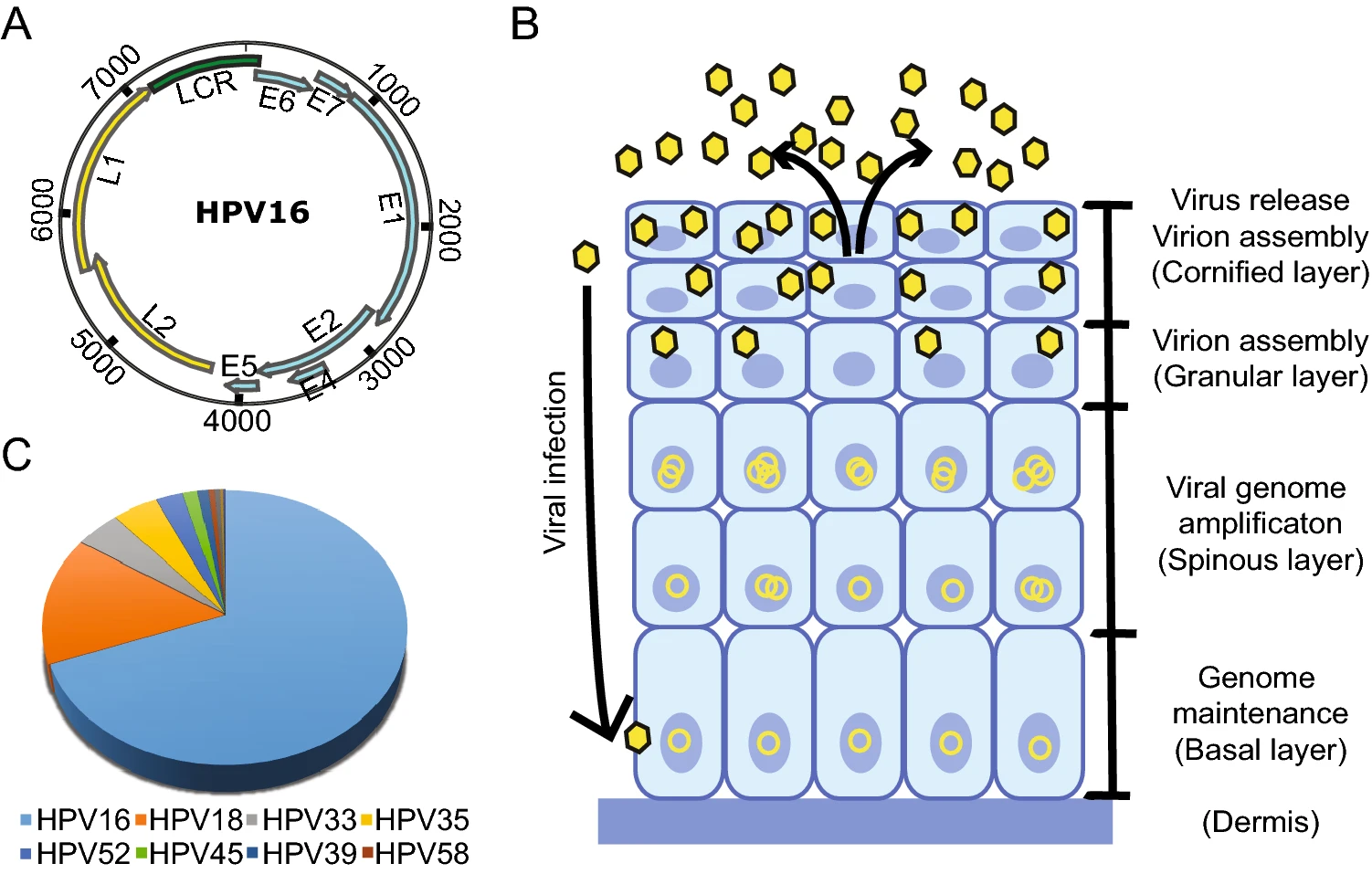
Current Status of Human Papillomavirus-Related Head and Neck Cancer: From Viral Genome to Patient Care
2021, 36(6): 1284 doi: 10.1007/s12250-021-00413-8
Human papillomavirus (HPV) infection identified as a definitive human carcinogen is increasingly being recognized for its role in carcinogenesis of human cancers. Up to 38%–80% of head and neck squamous cell carcinoma (HNSCC) in oropharyngeal location (OPSCC) and nearly all cervical cancers contain the HPV genome which is implicated in causing cancer through its oncoproteins E6 and E7. Given by the biologically distinct HPV-related OPSCC and a more favorable prognosis compared to HPV-negative tumors, clinical trials on de-escalation treatment strategies for these patients have been studied. It is therefore raised the questions for the patient stratification if treatment de-escalation is feasible. Moreover, understanding the crosstalk of HPV-mediated malignancy and immunity with clinical insights from the proportional response rate to immune checkpoint blockade treatments in patients with HNSCC is of importance to substantially improve the treatment efficacy. This review discusses the biology of HPV-related HNSCC as well as successful clinically findings with promising candidates in the pipeline for future directions. With the advent of various sequencing technologies, further biomolecules associated with HPV-related HNSCC progression are currently being identified to be used as potential biomarkers or targets for clinical decisions throughout the continuum of cancer care.
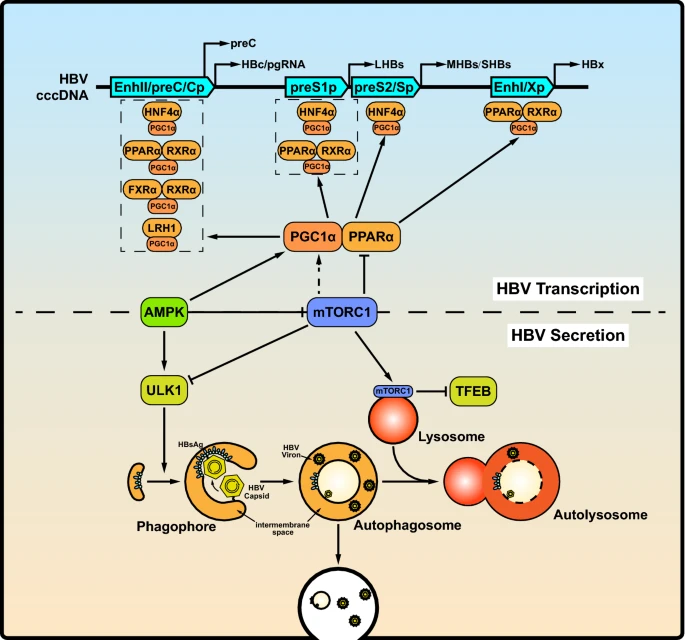
mTOR Signaling: The Interface Linking Cellular Metabolism and Hepatitis B Virus Replication
2021, 36(6): 1303 doi: 10.1007/s12250-021-00450-3
Mammalian target of rapamycin (mTOR) is a conserved Ser/Thr kinase that includes mTOR complex (mTORC) 1 and mTORC2. The mTOR pathway is activated in viral hepatitis, including hepatitis B virus (HBV) infection-induced hepatitis. Currently, chronic HBV infection remains one of the most serious public health issues worldwide. The unavailability of effective therapeutic strategies for HBV suggests that clarification of the pathogenesis of HBV infection is urgently required. Increasing evidence has shown that HBV infection can activate the mTOR pathway, indicating that HBV utilizes or hijacks the mTOR pathway to benefit its own replication. Therefore, the mTOR signaling pathway might be a crucial target for controlling HBV infection. Here, we summarize and discuss the latest findings from model biology research regarding the interaction between the mTOR signaling pathway and HBV replication.
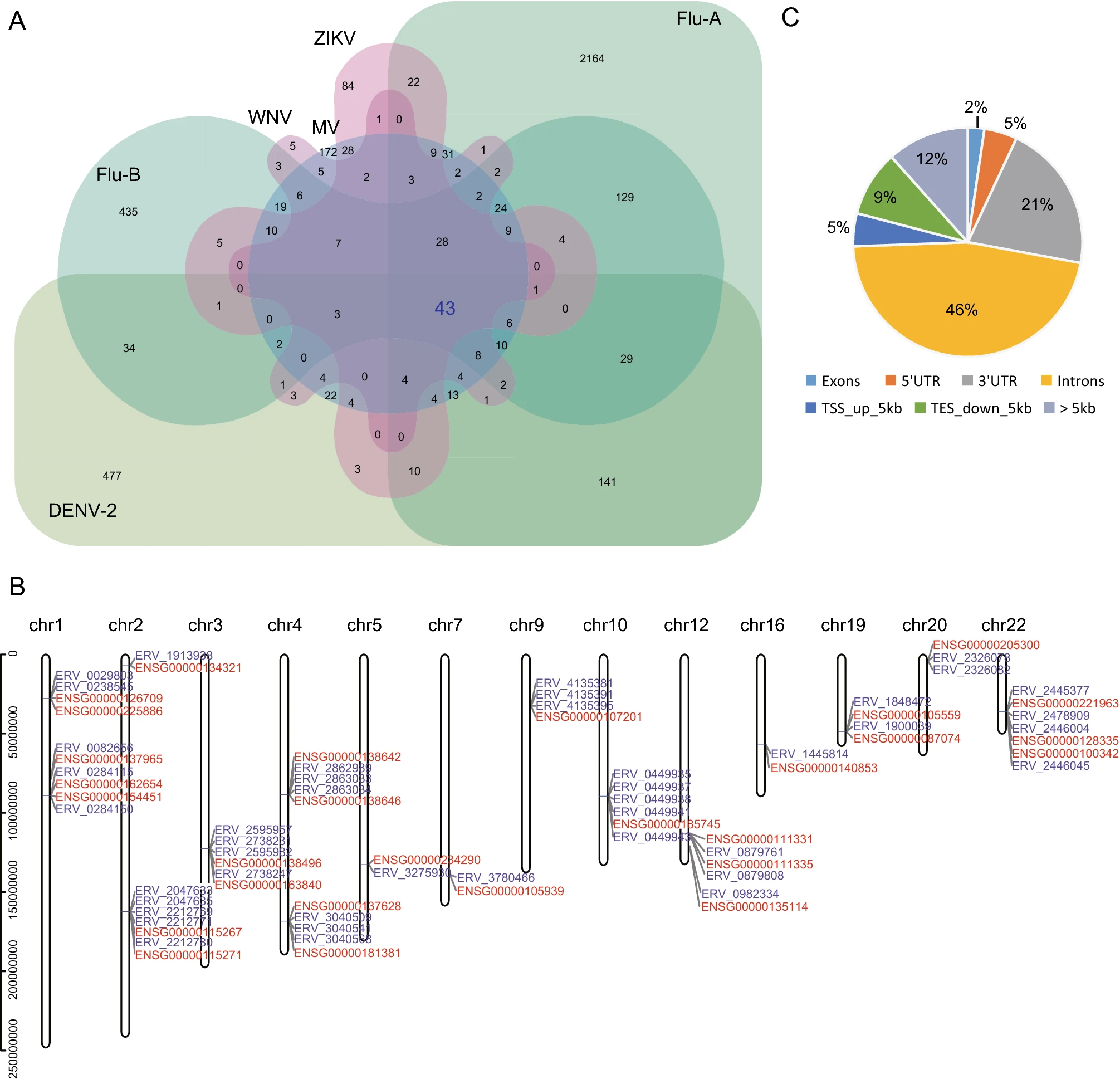
Transcriptome Analyses Implicate Endogenous Retroviruses Involved in the Host Antiviral Immune System through the Interferon Pathway
2021, 36(6): 1315 doi: 10.1007/s12250-021-00370-2
Human endogenous retroviruses (HERVs) are the remains of ancient retroviruses that invaded our ancestors' germline cell and were integrated into the genome. The expression of HERVs has always been a cause for concern because of its association with various cancers and diseases. However, few previous studies have focused on specific activation of HERVs by viral infections. Our previous study has shown that dengue virus type 2 (DENV-2) infection induces the transcription of a large number of abnormal HERVs loci; therefore, the purpose of this study was to explore the relationship between exogenous viral infection and HERV activation further. In this study, we retrieved and reanalyzed published data on 21 transcriptomes of human cells infected with various viruses. We found that infection with different viruses could induce transcriptional activation of HERV loci. Through the comparative analysis of all viral datasets, we identified 43 key HERV loci that were up-regulated by DENV-2, influenza A virus, influenza B virus, Zika virus, measles virus, and West Nile virus infections. Furthermore, the neighboring genes of these HERVs were simultaneously up-regulated, and almost all such neighboring genes were interferon-stimulated genes (ISGs), which are enriched in the host's antiviral immune response pathways. Our data supported the hypothesis that activation of HERVs, probably via an interferon-mediated mechanism, plays an important role in innate immunity against viral infections.
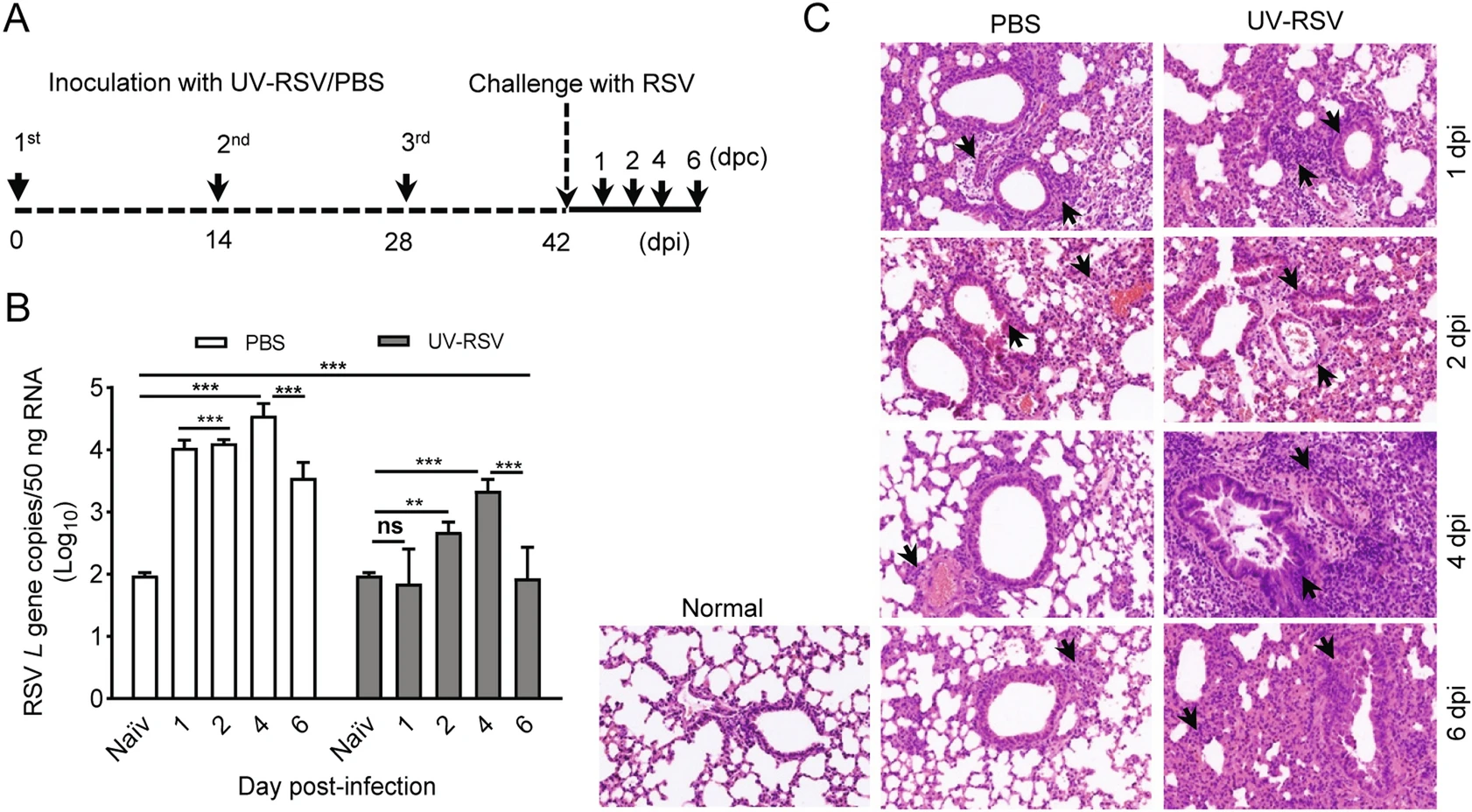
Dynamic Host Immune and Transcriptomic Responses to Respiratory Syncytial Virus Infection in a Vaccination-Challenge Mouse Model
2021, 36(6): 1327 doi: 10.1007/s12250-021-00418-3
Respiratory syncytial virus (RSV) is the major cause of lower respiratory tract infections in children. Inactivated RSV vaccine was developed in the late 1960's, but the vaccine-enhanced disease (VED) occurred to vaccinated infants upon subsequent natural RSV infection. The excessive inflammatory immunopathology in the lungs might be involved in the VED, but the underlying mechanisms remain not fully understood. In this study, we utilized UV-inactivated RSV in the prime/boost approach followed by RSV challenge in BALB/c mice to mimic RSV VED. The dynamic virus load, cytokines, histology and transcriptome profiles in lung tissues of mice were investigated from day 1 to day 6 post-infection. Compared to PBS-treated mice, UV-RSV vaccination leads to a Th2 type inflammatory response characterized by enhanced histopathology, reduced Treg cells and increased IL4+CD4 T cells in the lung. Enhanced production of several Th2 type cytokines (IL-4, IL-5, IL-10) and TGF-β, reduction of IL-6 and IL-17 were observed in UV-RSV vaccinated mice. A total of 5582 differentially expressed (DE) genes between PBS-treated or vaccinated mice and naïve mice were identified by RNA-Seq. Eleven conserved high-influential modules (HMs) were recognized, majorly grouped into regulatory networks related to cell cycle and cell metabolism, signal transduction, immune and inflammatory responses. At an early time post-infection, the vaccinated mice showed obvious decreased expression patterns of DE genes in 11 HMs compared to PBS-treated mice. The extracellular matrix (HM5) and immune responses (HM8) revealed tremendous differences in expression and regulation characteristics of transcripts between PBS-treated and vaccinated mice at both early and late time points. The highly connected genes in HM5 and HM8 networks were further validated by RT-qPCR. These findings reveal the relationship between RSV VED and immune responses, which could benefit the development of novel RSV vaccines.
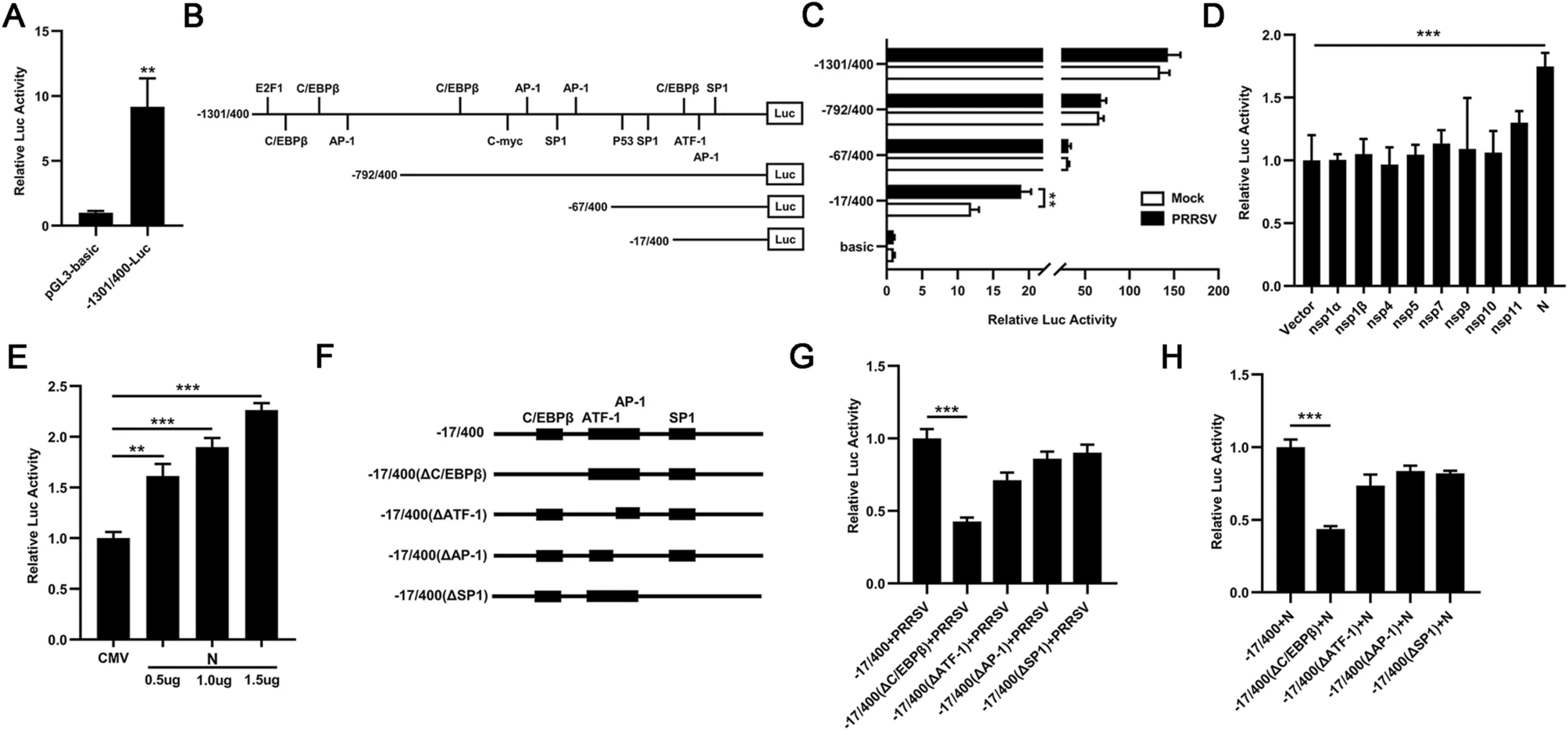
The C/EBPβ-Dependent Induction of TFDP2 Facilitates Porcine Reproductive and Respiratory Syndrome Virus Proliferation
2021, 36(6): 1341 doi: 10.1007/s12250-021-00403-w
Porcine reproductive and respiratory syndrome (PRRS) is an important infectious disease caused by porcine reproductive and respiratory syndrome virus (PRRSV), leading to significant economic losses in swine industry worldwide. Although several studies have shown that PRRSV can affect the cell cycle of infected cells, it is still unclear how it manipulates the cell cycle to facilitate its proliferation. In this study, we analyzed the mRNA expression profiles of transcription factors in PRRSV-infected 3D4/21 cells by RNA-sequencing. The result shows that the expression of transcription factor DP2 (TFDP2) is remarkably upregulated in PRRSV-infected cells. Further studies show that TFDP2 contributes to PRRSV proliferation and the PRRSV nucleocapsid (N) protein induces TFDP2 expression by activating C/EBPβ. TFDP2 positively regulates cyclin A expression and triggers a less proportion of cells in the S phase, which contributes to PRRSV proliferation. This study proposes a novel mechanism by which PRRSV utilizes host protein to regulate the cell cycle to favor its infection. Findings from this study will help us for a better understanding of PRRSV pathogenesis.
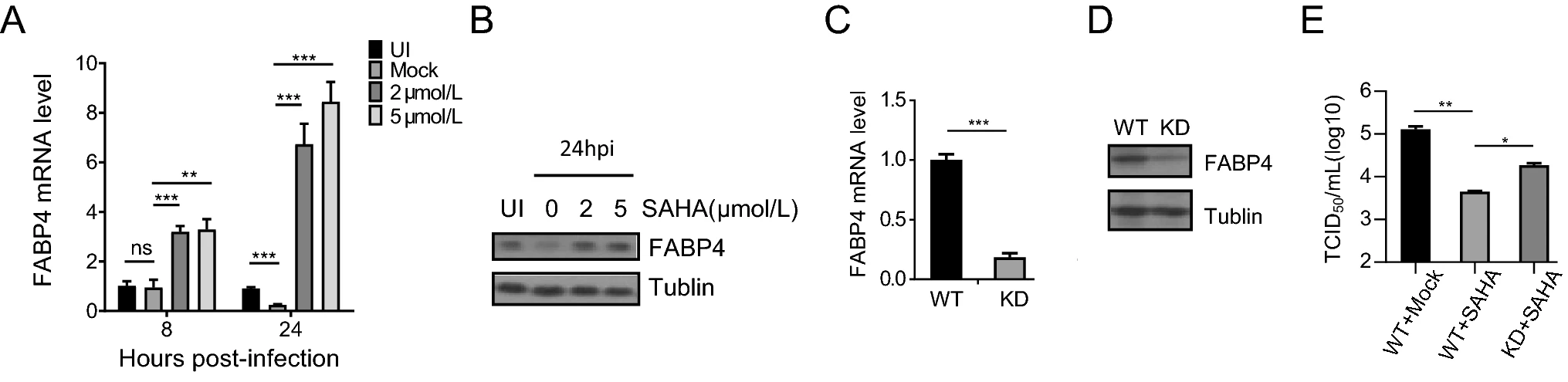
Histone Deacetylase Inhibitor SAHA Induces Expression of Fatty Acid-Binding Protein 4 and Inhibits Replication of Human Cytomegalovirus
2021, 36(6): 1352 doi: 10.1007/s12250-021-00382-y
Suberoylanilide hydroxamic acid (SAHA) is a histone deacetylase inhibitor that shows marked efficacy against many types of cancers and is approved to treat severe metastatic cutaneous T-cell lymphomas. In addition to its anticancer activity, SAHA has significant effects on the growth of many viruses. The effect of SAHA on replication of human cytomegalovirus (HCMV) has not, however, been investigated. Here, we showed that the replication of HCMV was significantly suppressed by treatment with SAHA at concentrations that did not show appreciable cytotoxicity. SAHA reduced transcription and protein levels of HCMV immediate early genes, showing that SAHA acts at an early stage in the viral life-cycle. RNA-sequencing data mining showed that numerous pathways and molecules were affected by SAHA. Interferon-mediated immunity was one of the most relevant pathways in the RNA-sequencing data, and we confirmed that SAHA inhibits HCMV-induced IFN-mediated immune responses using quantitative Real-time PCR (qRT-PCR). Fatty acid-binding protein 4 (FABP4), which plays a role in lipid metabolism, was identified by RNA-sequencing. We found that FABP4 expression was reduced by HCMV infection but increased by treatment with SAHA. We then showed that knockdown of FABP4 partially rescued the effect of SAHA on HCMV replication. Our data suggest that FABP4 contributes to the inhibitory effect of SAHA on HCMV replication.
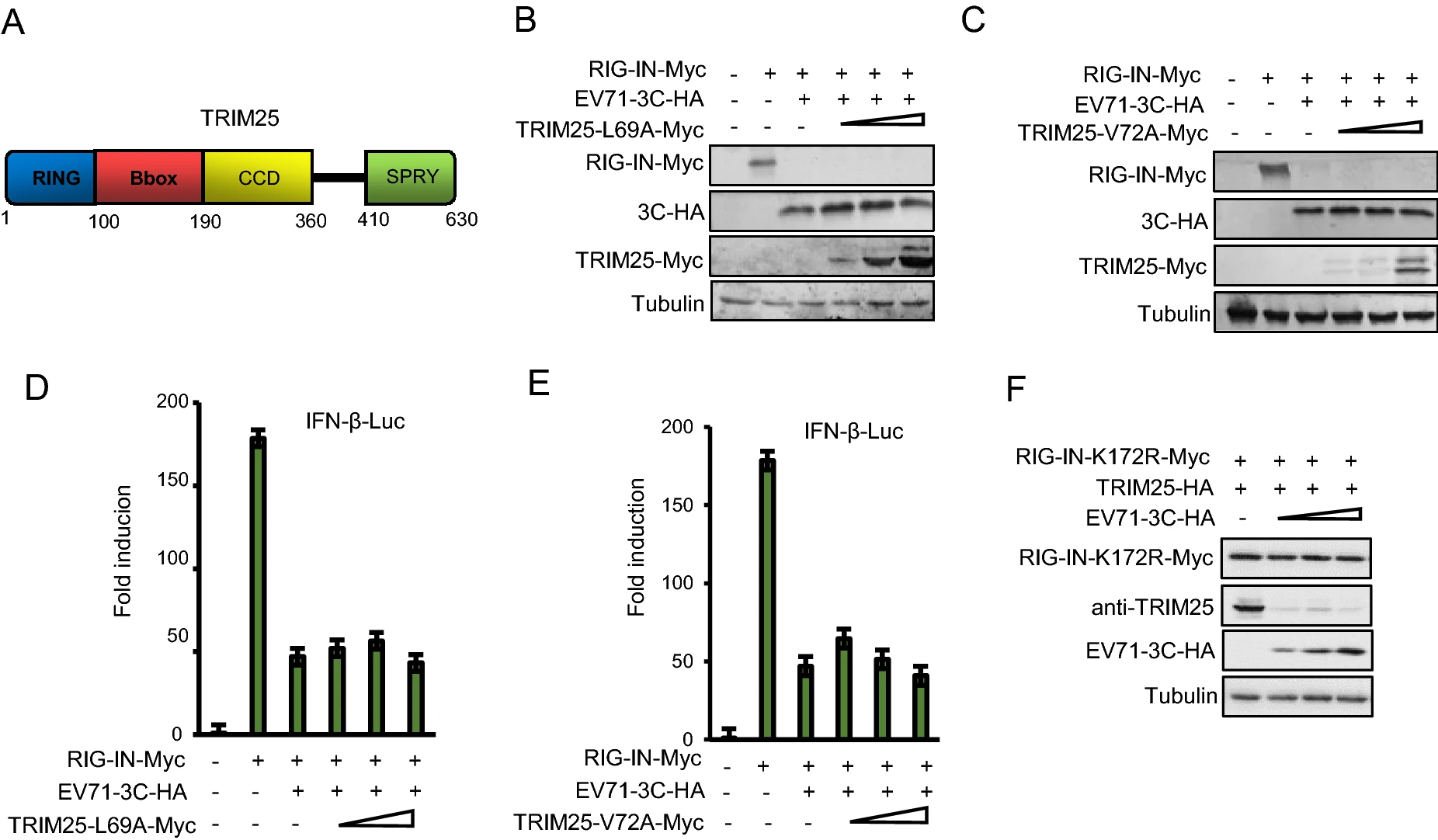
Ectopic Expression of TRIM25 Restores RIG-I Expression and IFN Production Reduced by Multiple Enteroviruses 3Cpro
2021, 36(6): 1363 doi: 10.1007/s12250-021-00410-x
Enteroviruses (EVs) 3C proteins suppress type Ⅰ interferon (IFN) responses mediated by retinoid acid-inducible gene I (RIG-I), while an E3 ubiquitin ligase, tripartite motif protein 25 (TRIM25)-mediated RIG-I ubiquitination is essential for RIG-I antiviral activity. Therefore, whether the effect of EVs 3C on RIG-I is associated with TRIM25 expression is worth to be further investigated. Here, we demonstrate that 3C proteins of EV71 and coxsackievirus B3 (CVB3) reduced not only RIG-I expression but also TRIM25 expression through protease cleavage activity, while overexpression of TRIM25 restored RIG-I expression and IFN-β production reduced by 3C proteins. Further investigation confirmed that the two amino acids and functional domains in TRIM25 required for RIG-I ubiquitination and TRIM25 structural conformation were essential for the recovery of RIG-I expression. Moreover, we also observed that TRIM25 could rescue RIG-I expression reduced by 3C proteins of CVA6 and EV-D68 but not CVA16. Our findings provide an insightful interpretation of 3C-mediated host innate immune suppression and support TRIM25 as an attractive target against multiple EVs infection.
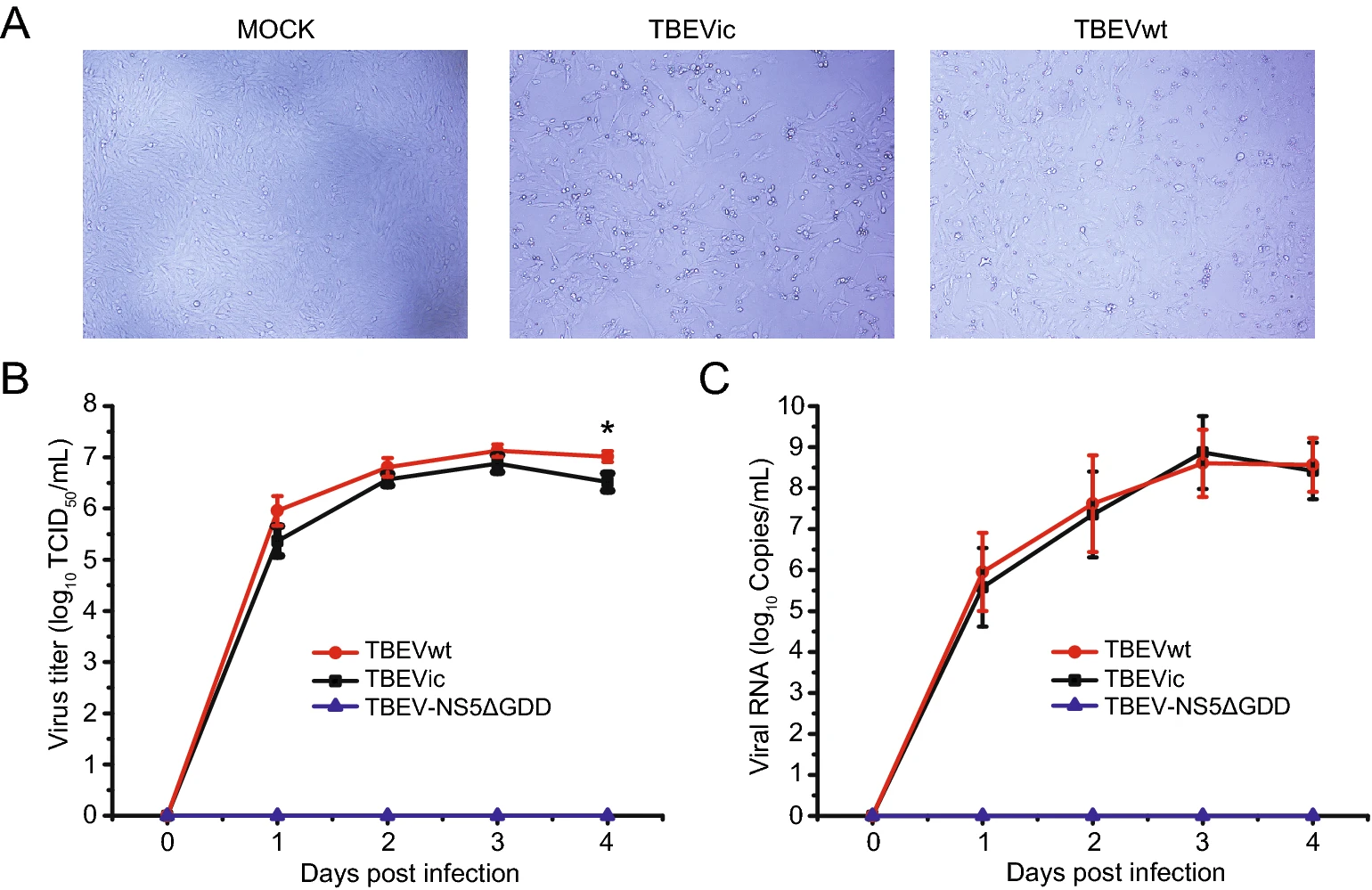
Recovery of a Far-Eastern Strain of Tick-Borne Encephalitis Virus with a Full-Length Infectious cDNA Clone
2021, 36(6): 1375 doi: 10.1007/s12250-021-00396-6
Tick-borne encephalitis virus (TBEV) is a pathogenic virus known to cause central nervous system (CNS) diseases in humans, and has become an increasing public health threat nowadays. The rates of TBEV infection in the endemic countries are increasing. However, there is no effective antiviral against the disease. This underscores the urgent need for tools to study the emergence and pathogenesis of TBEV and to accelerate the development of vaccines and antivirals. In this study, we reported an infectious cDNA clone of TBEV that was isolated in China (the WH2012 strain). A beta-globin intron was inserted in the coding region of nonstructural protein 1 (NS1) gene to improve the stability of viral genome in bacteria. In mammalian cells, the inserted intron was excised and spliced precisely, which did not lead to the generation of inserted mutants. High titers of infectious progeny viruses were generated after the transfection of the infectious clone. The cDNA-derived TBEV replicated efficiently, and caused typical cytopathic effect (CPE) and plaques in BHK-21 cells. In addition, the CPE and growth curve of cDNA-derived virus were similar to that of its parental isolate in cells. Together, we have constructed the first infectious TBEV cDNA clone in China, and the clone can be used to investigate the genetic determinants of TBEV virulence and disease pathogenesis, and to develop countermeasures against the virus.
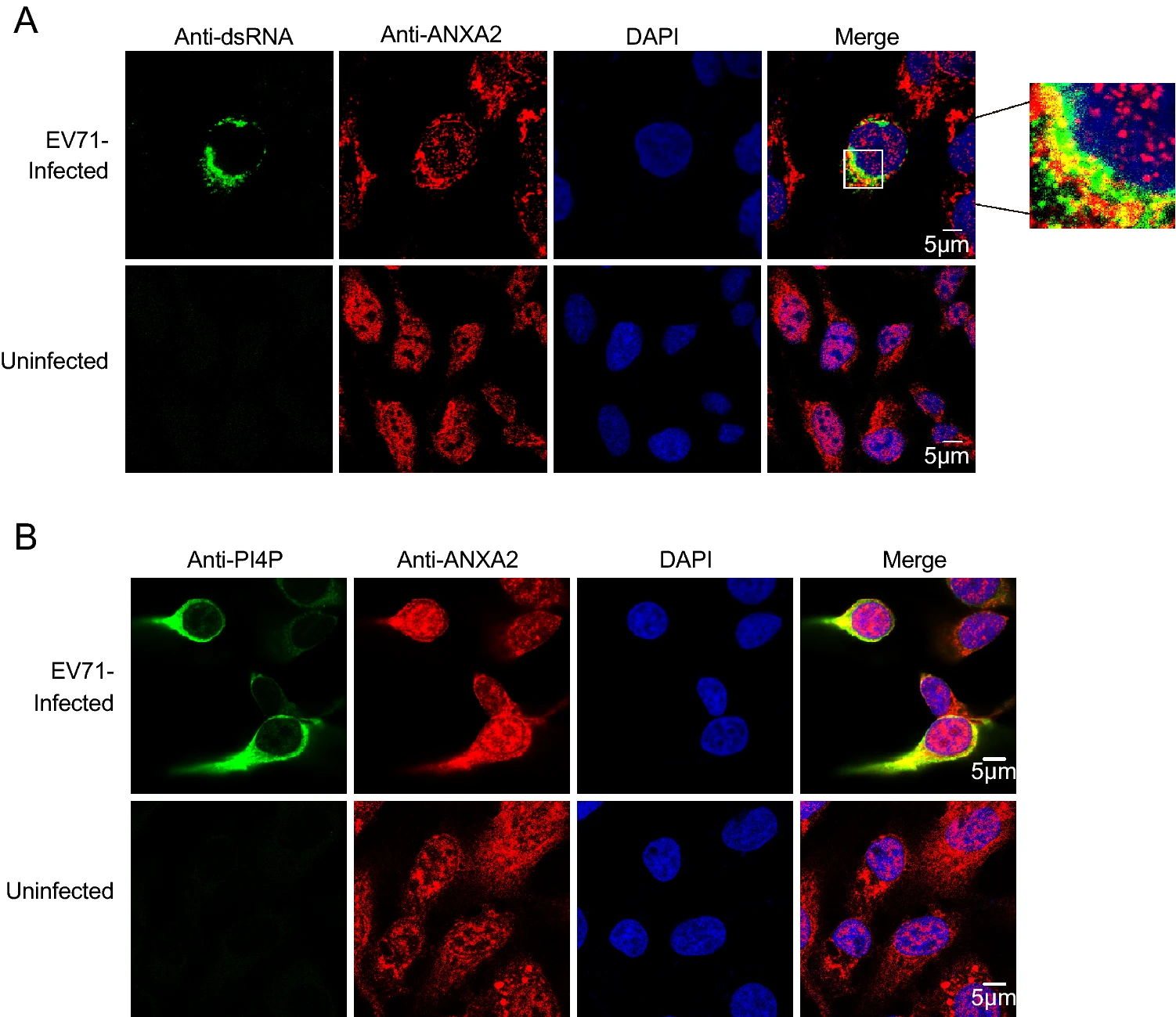
ANXA2 Facilitates Enterovirus 71 Infection by Interacting with 3D Polymerase and PI4KB to Assist the Assembly of Replication Organelles
2021, 36(6): 1387 doi: 10.1007/s12250-021-00417-4
Similar to that of other enteroviruses, the replication of enterovirus 71 (EV71) occurs on rearranged membranous structures called replication organelles (ROs). Phosphatidylinositol 4-kinase Ⅲ (PI4KB), which is required by enteroviruses for RO formation, yields phosphatidylinositol-4-phosphate (PI4P) on ROs. PI4P then binds and induces conformational changes in the RNA-dependent RNA polymerase (RdRp) to modulate RdRp activity. Here, we targeted 3D polymerase, the core enzyme of EV71 ROs, and found that the host factor Annexin A2 (ANXA2) can interact with 3D polymerase and promote the replication of EV71. Then, an experiment showed that the annexin domain of ANXA2, which possesses membrane-binding capacity, mediates the interaction of ANXA2 with EV71 3D polymerase. Further research showed that ANXA2 is localized on ROs and interacts with PI4KB. Overexpression of ANXA2 stimulated the formation of PI4P, and the level of PI4P was decreased in ANXA2-knockout cells. Furthermore, ANXA2, PI4KB, and 3D were shown to be localized to the viral RNA replication site, where they form a higher-order protein complex, and the presence of ANXA2 promoted the PI4KB-3D interaction. Altogether, our data provide new insight into the role of ANXA2 in facilitating formation of the EV71 RNA replication complex.
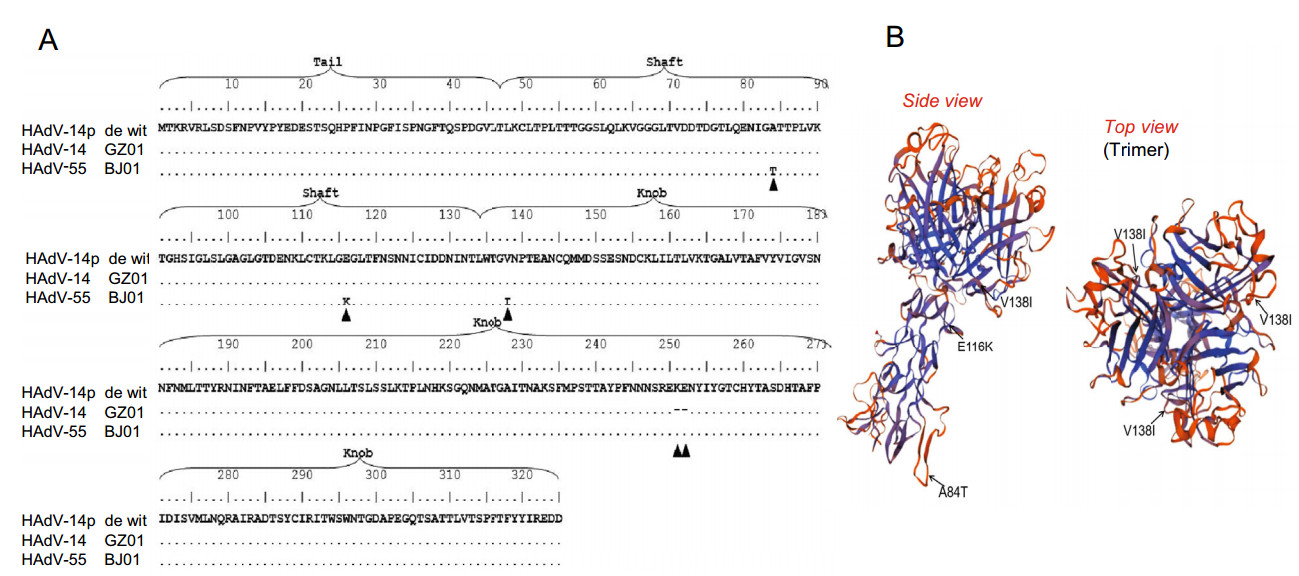
Desmoglein 2 (DSG2) Is A Receptor of Human Adenovirus Type 55 Causing Adult Severe Community-Acquired Pneumonia
2021, 36(6): 1400 doi: 10.1007/s12250-021-00414-7
Human adenovirus type 55 (HAdV-B55) is a re-emergent acute respiratory disease pathogen that causes adult community-acquired pneumonia (CAP). Previous studies have shown that the receptor of HAdV-B14, which genome is highly similar with HAdV-B55, is human Desmoglein 2 (DSG2). However, whether the receptor of HAdV-B55 is DSG2 is undetermined because there are three amino acid mutations in the fiber gene between HAdV-B14 and HAdV-B55. Here, firstly we found the 3T3 cells, a mouse embryo fibroblast rodent cell line which does not express human DSG2, were able to be infected by HAdV-B55 after transfected with pcDNA3.1-DSG2, while normal 3T3 cells were still unsusceptible to HAdV-B55 infection. Next, A549 cells with hDSG2 knock-down by siRNA were hard to be infected by HAdV-B3/-B14/-B55, while the control siRNA group was still able to be infected by all these types of HAdVs. Finally, immunofluorescence confocal microscopy indicated visually that Cy3-conjugated HAdV-B55 viruses entered A549 cells by binding to DSG2 protein. Therefore, DSG2 is a major receptor of HAdV-B55 causing adult CAP. Our finding is important for better understanding of interactions between adenoviruses and host cells and may shed light on the development of new drugs that can interfere with these processes as well as for the development of potent prophylactic vaccines.

Non-Structural Protein 5 of Zika Virus Interacts with p53 in Human Neural Progenitor Cells and Induces p53-Mediated Apoptosis
2021, 36(6): 1411 doi: 10.1007/s12250-021-00422-7
Zika virus (ZIKV) infection could disrupt neurogenesis and cause microcephaly in neonates by targeting neural progenitor cells (NPCs). The tumor suppressor p53-mediated cell cycle arrest and apoptotic cell death have been suggested to be activated upon ZIKV infection, yet the detailed mechanism is not well understood. In the present study, we investigated the effects of ZIKV-encoded proteins in the activation of p53 signaling pathway and found that, among the ten viral proteins, the nonstructural protein 5 (NS5) of ZIKV most significantly activated the transcription of p53 target genes. Using the immunoprecipitation-coupled mass spectrometry approach, we identified that ZIKV-NS5 interacted with p53 protein. The NS5-p53 interaction was further confirmed by co-immunoprecipitation and GST pull-down assays. In addition, the MTase domain of NS5 and the C-terminal domain of p53 were mapped to be responsible for the interaction between these two proteins. We further showed that ZIKV-NS5 was colocalized with p53 and increased its protein level in the nuclei and able to prolong the half-life of p53. Furthermore, lentivirus-mediated expression of ZIKV-NS5 in hNPCs led to an apparent cell death phenotype. ZIKV-NS5 promoted the cleavage of PARP1 and significantly increased the cell apoptosis of hNPCs. Taken together, these findings revealed that ZIKV-NS5 is a previously undiscovered regulator of p53-mediated apoptosis in hNPCs, which may contribute to the ZIKV-caused abnormal neurodevelopment.
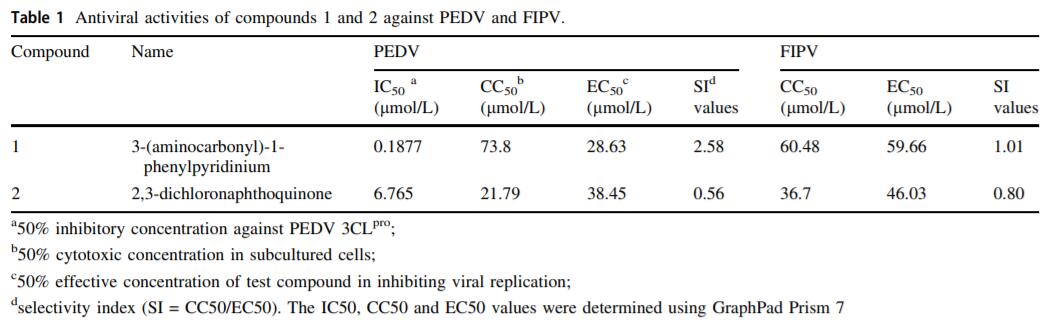
Two Inhibitors Against the 3C-Like Proteases of Swine Coronavirus and Feline Coronavirus
2021, 36(6): 1421 doi: 10.1007/s12250-021-00415-6
Coronaviruses (CoVs) are important human and animal pathogens that cause respiratory and gastrointestinal diseases. Porcine epidemic diarrhoea (PED), characterized by severe diarrhoea and vomiting in pigs, is a highly lethal disease caused by porcine epidemic diarrhoea virus (PEDV) and causes substantial losses in the swine industry worldwide. However, currently available commercial drugs have not shown great therapeutic effects. In this study, a fluorescence resonance energy transfer (FRET)-based assay was applied to screen a library containing 1, 590 compounds and identified two compounds, 3-(aminocarbonyl)-1-phenylpyridinium and 2, 3-dichloronaphthoquinone, that target the 3C-like protease (3CLpro) of PEDV. These compounds are of low molecular weight (MW) and greatly inhibited the activity of this enzyme (IC50 values were obtained in this study). Furthermore, these compounds exhibited antiviral capacity against another member of the CoV family, feline infectious peritonitis virus (FIPV). Here, the inhibitory effects of these compounds against CoVs on Vero cells and feline kidney cells were identified (with EC50 values) and cell viability assays were performed. The results of putative molecular docking models indicate that these compounds, labeled compound 1 and compound 2, contact the conserved active sites (Cys144, Glu165, Gln191) of 3CLpro via hydrogen bonds. These findings provide insight into the antiviral activities of compounds 1 and 2 that may facilitate future research on anti-CoV drugs.
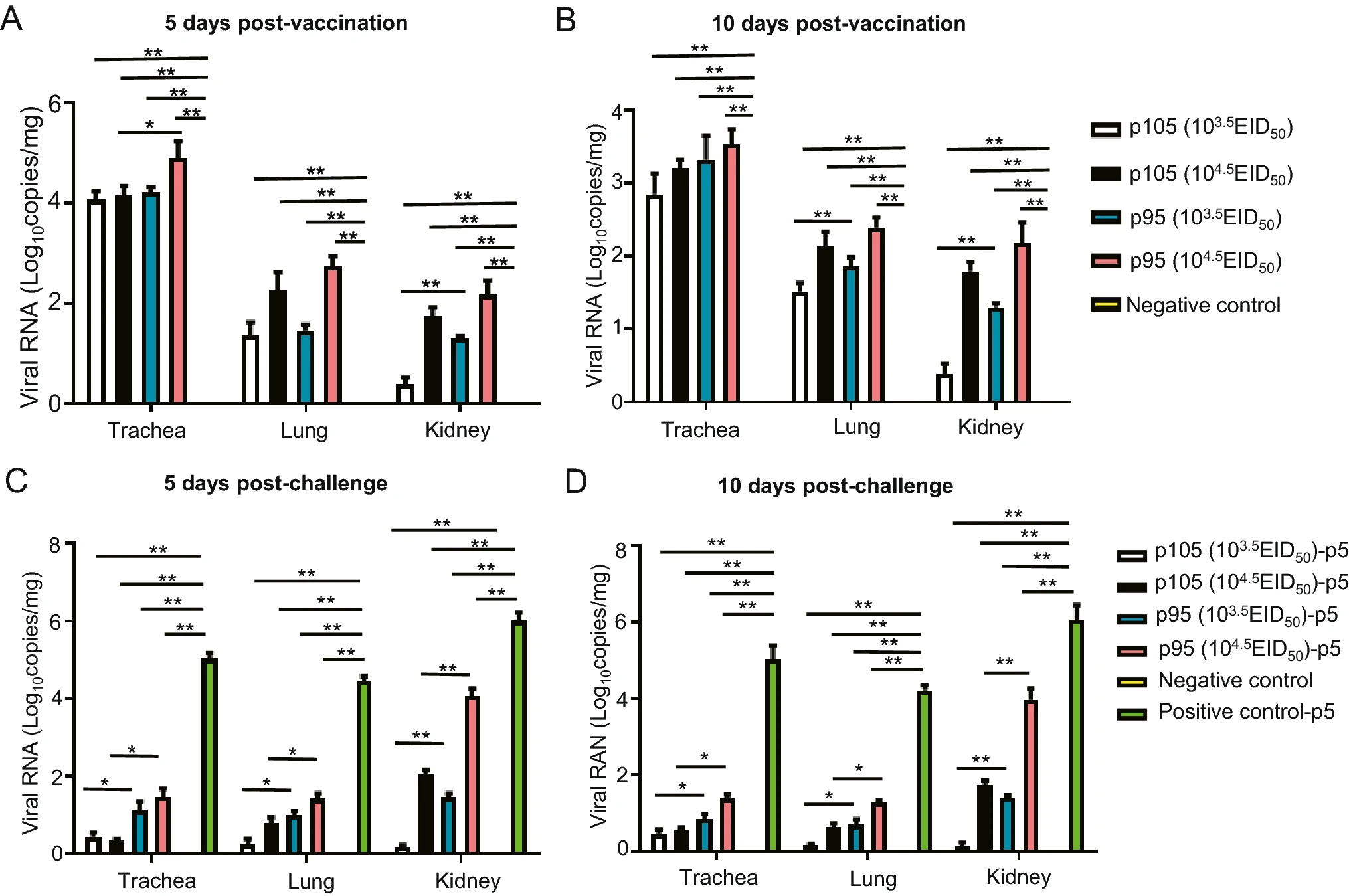
The Efficacy of a Live Attenuated TW I-Type Infectious Bronchitis Virus Vaccine Candidate
2021, 36(6): 1431 doi: 10.1007/s12250-021-00419-2
Infectious bronchitis (IB) is a highly contagious avian disease caused by infection with infectious bronchitis virus (IBV), which seriously affects the development of the global poultry industry. The distribution of TW I-type IBV in China has increased in recent years, becoming a widespread genotype. We previously isolated a TW I-type IBV strain termed CK/CH/GD/GZ14 in 2014, but its pathogenicity and possibility for vaccine development were not explored. Therefore, this research aimed to develop a live-attenuated virus vaccine based on the CK/CH/GD/GZ14 strain. The wild type IBV CK/CH/GD/GZ14 strain was serially passaged in SPF embryos for 145 generations. The morbidity and mortality rate of wild-type strain in 14 day-old chickens is 100% and 80% respectively, while the morbidity rate in the attenuated strain was 20% in the 95th and 105th generations and there was no death. Histopathological observations showed that the pathogenicity of the 95th and 105th generations in chickens was significantly weakened. Further challenge experiments confirmed that the attenuated CK/CH/GD/GZ14 strain in the 95th and 105th generations could resist CK/CH/GD/GZ14 (5th generation) infection and the protection rate was 80%. Tracheal cilia stagnation, virus shedding, and viral load experiments confirmed that the 95th and 105th generations provide good immune protection in chickens, and the immunogenicity of the 105th generation is better than that of the 95th generation. These data suggest that the attenuated CK/CH/GD/GZ14 strain in the 105th generation may be applied as a vaccine candidate against TW I-type IBV.
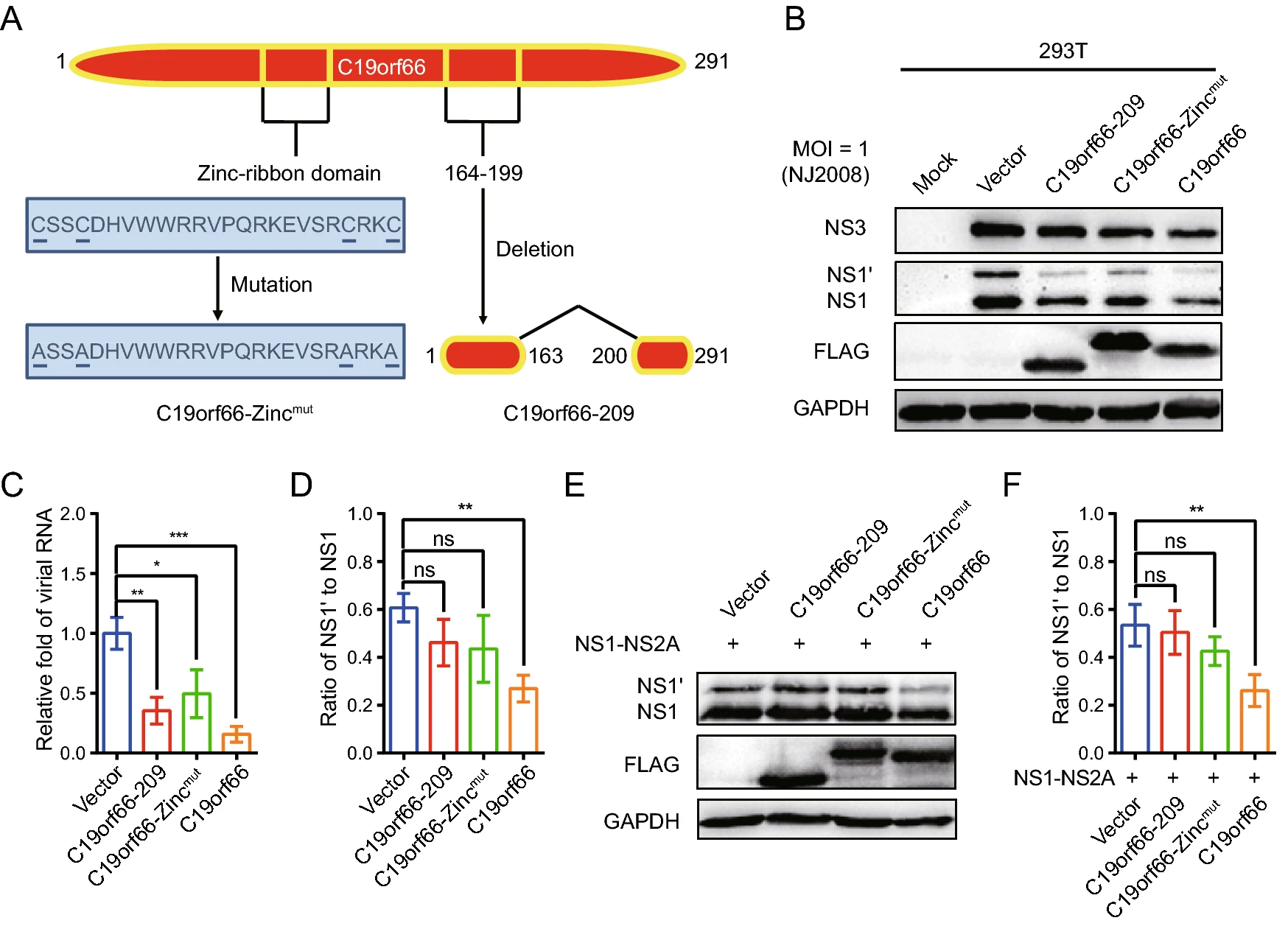
C19orf66 Inhibits Japanese Encephalitis Virus Replication by Targeting -1 PRF and the NS3 Protein
2021, 36(6): 1443 doi: 10.1007/s12250-021-00423-6
The Japanese encephalitis serogroup of the neurogenic Flavivirus has a specific feature that expresses a non-structural protein NS1' produced through a programmed -1 ribosomal frameshifting (-1 PRF). Herein, C19orf66, a novel member of interferon-stimulated gene (ISG) products, exhibited significant activity of antagonizing Japanese encephalitis virus (JEV) infection. Overexpression of C19orf66 in 293T cells significantly inhibited JEV replication, while knock-down of endogenous C19orf66 in HeLa cells and A549 cells significantly increased virus replication. Notably, C19orf66 had an inhibitory effect on frameshift production of JEV NS1'. The inhibition was more significant when C19orf66 and JEV NS1-NS2A were co-expressed in the 293T cells. Both C19orf66-209 and C19orf66-Zincmut did not significantly change the NS1' to NS1 ratio and had weaker antiviral effects than C19orf66. Similarly, C19orf66-209 and C19orf66-Zincmut had no significant effect on the expression of the JEV NS3 protein, whose expression was down-regulated by C19orf66 via the lysosome-dependent pathway. These findings suggest that C19orf66 may possess at least two different mechanisms of antagonizing JEV infection. This study identified C19orf66 as a novel interferon-stimulated gene product that can inhibit JEV replication by targeting -1 PRF and the NS3 protein. The study provides baseline information for the future devel-opment of broad-spectrum antiviral agents against JEV.
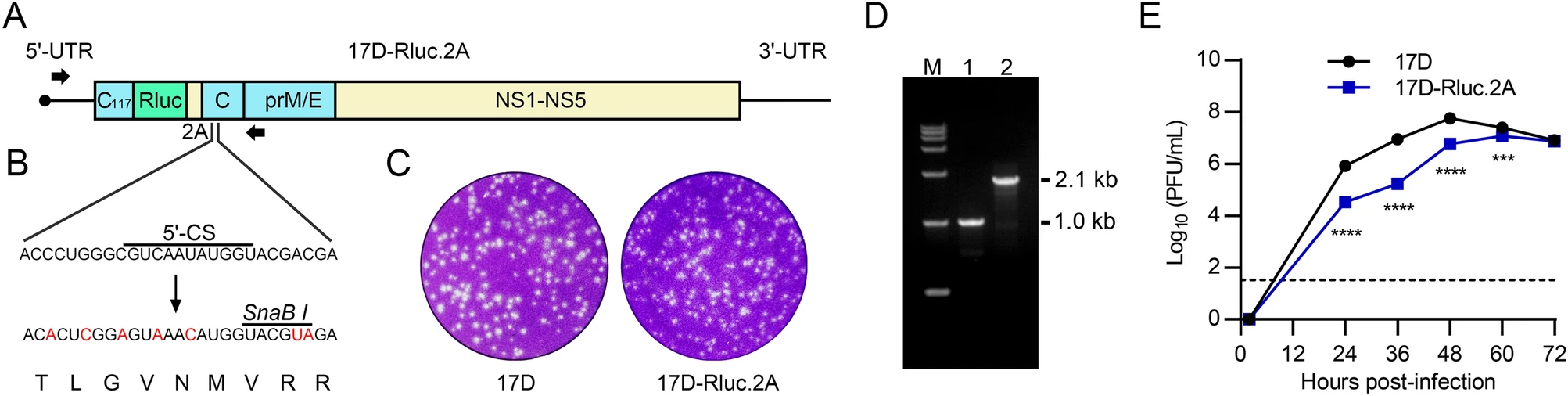
Generation and Application of a Luciferase Reporter Virus Based on Yellow Fever Virus 17D
2021, 36(6): 1456 doi: 10.1007/s12250-021-00428-1
Yellow fever virus (YFV) is a re-emerging virus that can cause life-threatening yellow fever disease in humans. Despite the availability of an effective vaccine, little is known about the replication mechanism of YFV, and there are still no available specific anti-YFV medicines. Herein, by introducing the Renilla luciferase gene (Rluc) into an infectious clone of YFV vaccine strain 17D, we generated a recombinant virus 17D-Rluc.2A via reverse genetics approaches. The 17D-Rluc.2A had similar plaque morphology and comparable in vitro growth characteristics with its parental strain. Importantly, the reporter luciferase was efficiently expressed in 17D-Rluc.2A-infected mammalian and mosquito cells, and there was a good linear correlation between intracellular luciferase expression and extracellular infectious virion reproduction. Furthermore, by a combination of the 17D-Rluc.2A reporter virus and selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) technology, the conserved 5′-SLA element was shown to be essential for YFV replication, highlighting the capability of 17D-Rluc.2A in the investigation of YFV replication. At last, we demonstrated that two compounds with distinct anti-viral mechanisms can effectively inhibit the viral propagation in 17D-Rluc.2A-infected cells, demonstrating its potential application in the evaluation of anti-viral medicines. Taken together, the 17D-Rluc.2A serves as a useful tool for the study of YFV replication and anti-YFV medicine development.
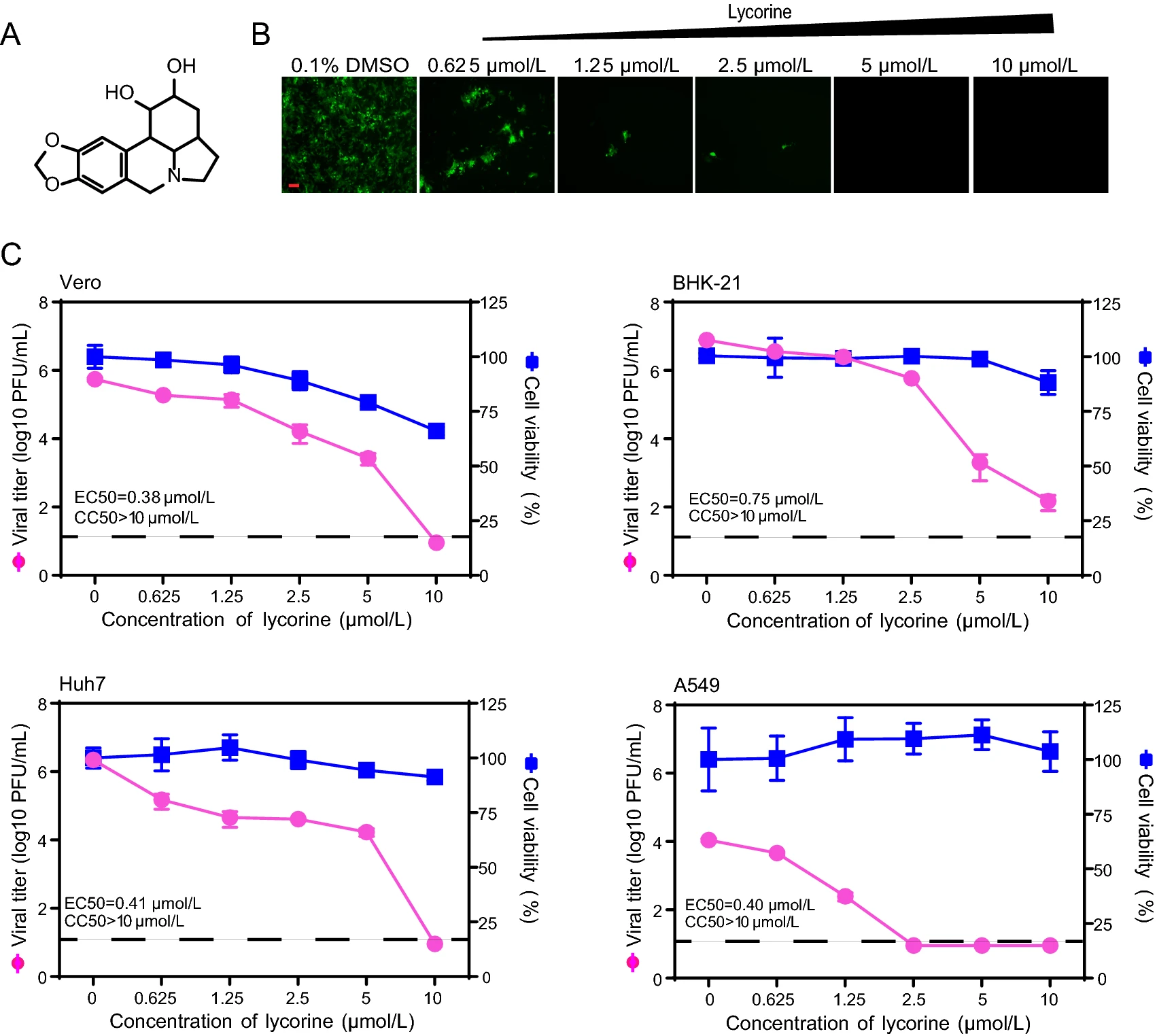
In Vitro Inhibition of Alphaviruses by Lycorine
2021, 36(6): 1465 doi: 10.1007/s12250-021-00438-z
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus. As an emerging virus, CHIKV imposes a threat to public health. Currently, there are no vaccines or antivirals available for the prevention of CHIKV infection. Lycorine, an alkaloid from Amaryllidaceae plants, has antiviral activity against a number of viruses such as coronavirus, flavivirus and enterovirus. In this study, we found that lycorine could inhibit CHIKV in cell culture at a concentration of 10 μmol/L without apparent cytotoxicity. In addition, it exhibited broad-spectrum anti-alphavirus activity, including Sindbis virus (SINV), Semliki Forest virus (SFV), and Venezuelan equine encephalomyelitis virus (VEEV). The time of addition studies indicated that lycorine functions at an early post-entry stage of CHIKV life cycle. The results based on two different CHIKV replicons provided further evidence that lycorine exerts its antiviral activity mainly by inhibiting CHIKV translation. Overall, our study extends the antiviral spectrum of lycorine.
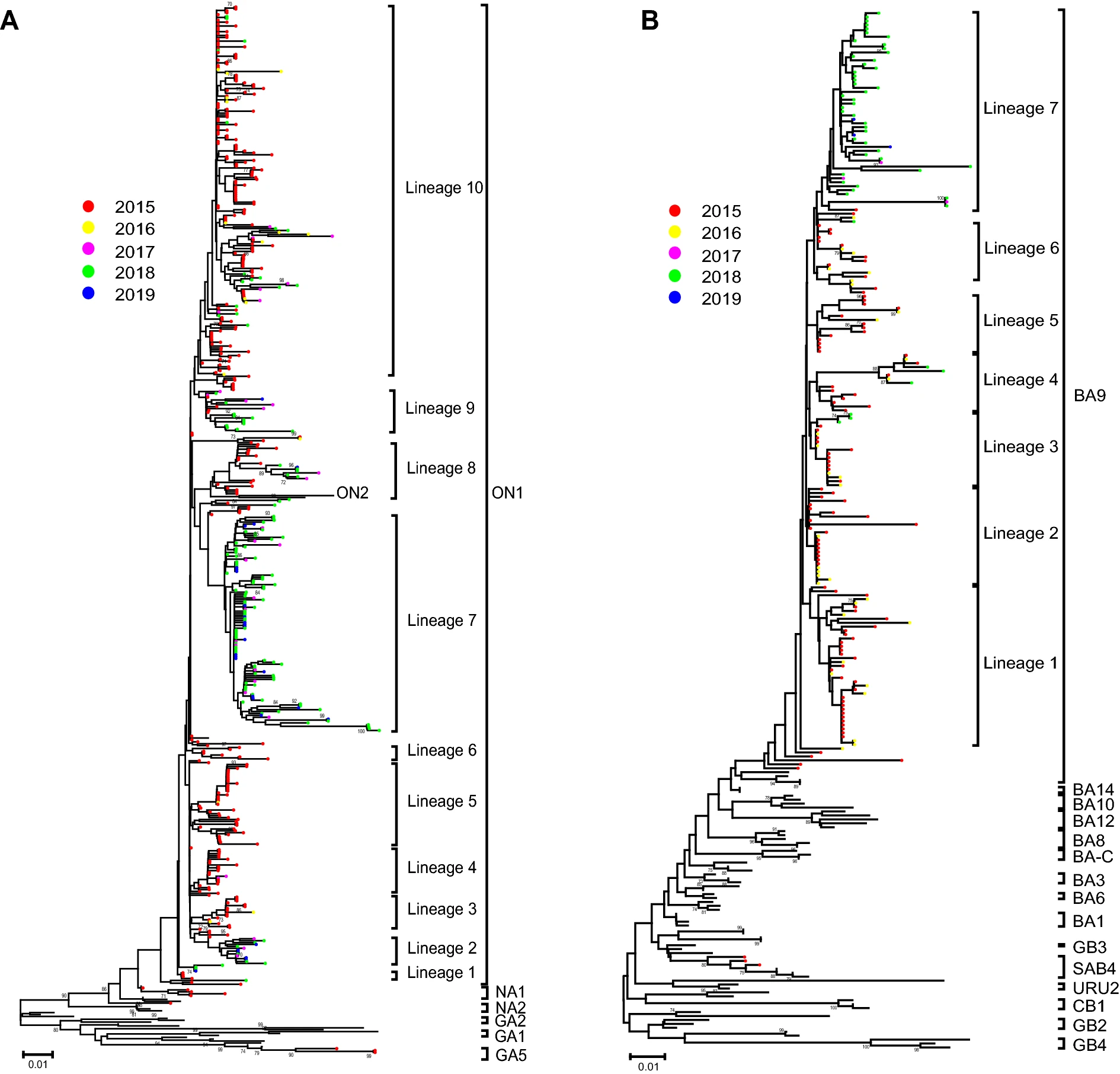
A multi-center study on Molecular Epidemiology of Human Respiratory Syncytial Virus from Children with Acute Lower Respiratory Tract Infections in the Mainland of China between 2015 and 2019
2021, 36(6): 1475 doi: 10.1007/s12250-021-00430-7
Human respiratory syncytial virus (RSV) is a major pathogen of acute lower respiratory tract infection among young children. To investigate the prevalence and genetic characteristics of RSV in China, we performed a molecular epidemiological study during 2015-2019. A total of 964 RSV-positive specimens were identified from 5529 enrolled patients during a multi-center study. RSV subgroup A (RSV-A) was the predominant subgroup during this research period except in 2016. Totally, 535 sequences of the second hypervariable region (HVR-2) of the G gene were obtained. Combined with 182 Chinese sequences from GenBank, phylogenetic trees showed that 521 RSV-A sequences fell in genotypes ON1 (512), NA1 (6) and GA5 (3), respectively; while 196 RSV-B sequences fell in BA9 (193) and SAB4 (3). ON1 and BA9 were the only genotypes after December 2015. Genotypes ON1 and BA9 can be separated into 10 and 7 lineages, respectively. The HVR-2 of genotype ON1 had six amino acid changes with a frequency more than 10%, while two substitutions H258Q and H266L were co-occurrences. The HVR-2 of genotype BA9 had nine amino acid substitutions with a frequency more than 10%, while the sequences with T290I and T312I were all from 2018 to 2019. One N-glycosylation site at 237 was identified among ON1 sequences, while two N-glycosylation sites (296 and 310) were identified in the 60-nucleotide duplication region of BA9. To conclusion, ON1 and BA9 were the predominant genotypes in China during 2015-2019. For the genotypes ON1 and BA9, the G gene exhibited relatively high diversity and evolved continuously.
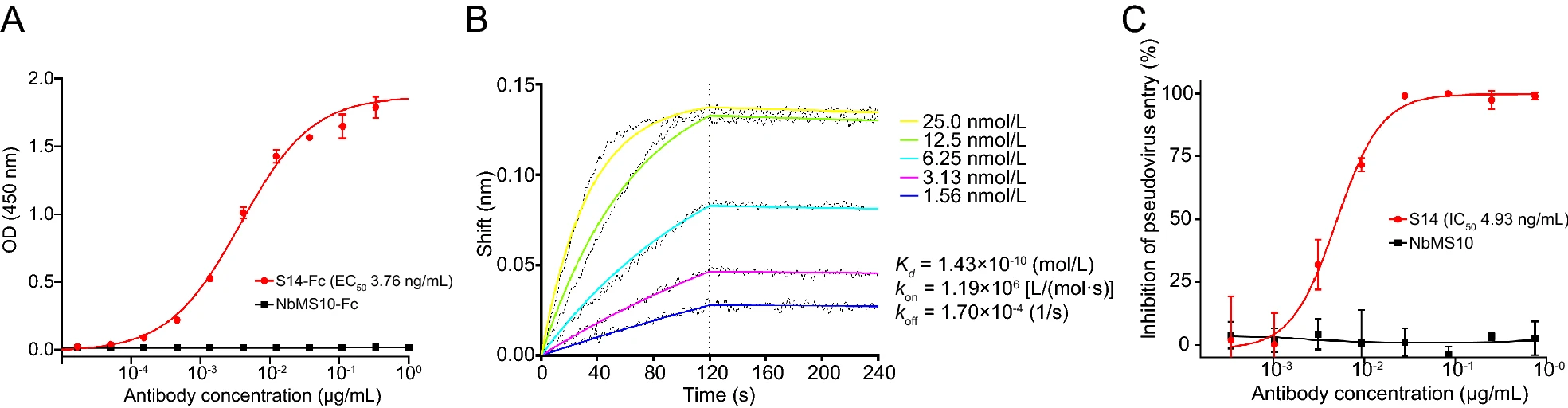
Generation and Characterization of a Nanobody Against SARS-CoV
2021, 36(6): 1484 doi: 10.1007/s12250-021-00436-1
The sudden emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) has caused global panic in 2003, and the risk of SARS-CoV outbreak still exists. However, no specific antiviral drug or vaccine is available; thus, the development of therapeutic antibodies against SARS-CoV is needed. In this study, a nanobody phage-displayed library was constructed from peripheral blood mononuclear cells of alpacas immunized with the recombinant receptor-binding domain (RBD) of SARS-CoV. Four positive clones were selected after four rounds of bio-panning and subjected to recombinant expression in E. coli. Further biological identification demonstrated that one of the nanobodies, S14, showed high affinity to SARS-CoV RBD and potent neutralization activity at the picomole level against SARS-CoV pseudovirus. A competitive inhibition assay showed that S14 blocked the binding of SARS-CoV RBD to either soluble or cell-expressed angiotensinconverting enzyme 2 (ACE2). In summary, we developed a novel nanobody targeting SARS-CoV RBD, which might be useful for the development of therapeutics against SARS.

Characterization of Episomal Replication of Bovine Papillomavirus Type 1 DNA in Long-Term Virion-Infected Saccharomyces Cerevisiae Culture
2021, 36(6): 1492 doi: 10.1007/s12250-021-00439-y
We have previously reported that bovine papillomavirus type 1 (BPV-1) DNA can replicate its genome and produce infectious virus-like particles in short term virion-infected S. cerevisiae (budding yeast) cultures (Zhao and Frazer 2002, Journal of Virology, 76:3359–64 and 76:12265–73). Here, we report the episomal replications of BPV-1 DNA in long term virion-infected S. cerevisiae culture up to 108 days. Episomal replications of the BPV-1 DNA could be divided into three patterns at three stages, early active replication (day 3–16), middle weak replication (day 23–34/45) and late stable replication (day 45–82). Two-dimensional gel electrophoresis analysis and Southern blot hybridization have revealed further that multiple replication intermediates of BPV-1 DNA including linear form, stranded DNA, monomers and higher oligomers were detected in the virion-infected yeast cells over the time course. Higher oligomers shown as covalently closed circular DNAs (cccDNAs) are the most important replication intermediates that serve as the main nuclear transcription template for producing all viral RNAs in the viral life cycle. In this study, the cccDNAs were generated at the early active replication stage with the highest frequencies and then at late stable replication, but they appeared to be suppressed at the middle weak replication. Our data provided a novel insight that BPV-1 genomic DNA could replicate episomally for the long period and produce the key replication intermediates cccDNAs in S. cerevisiae system.
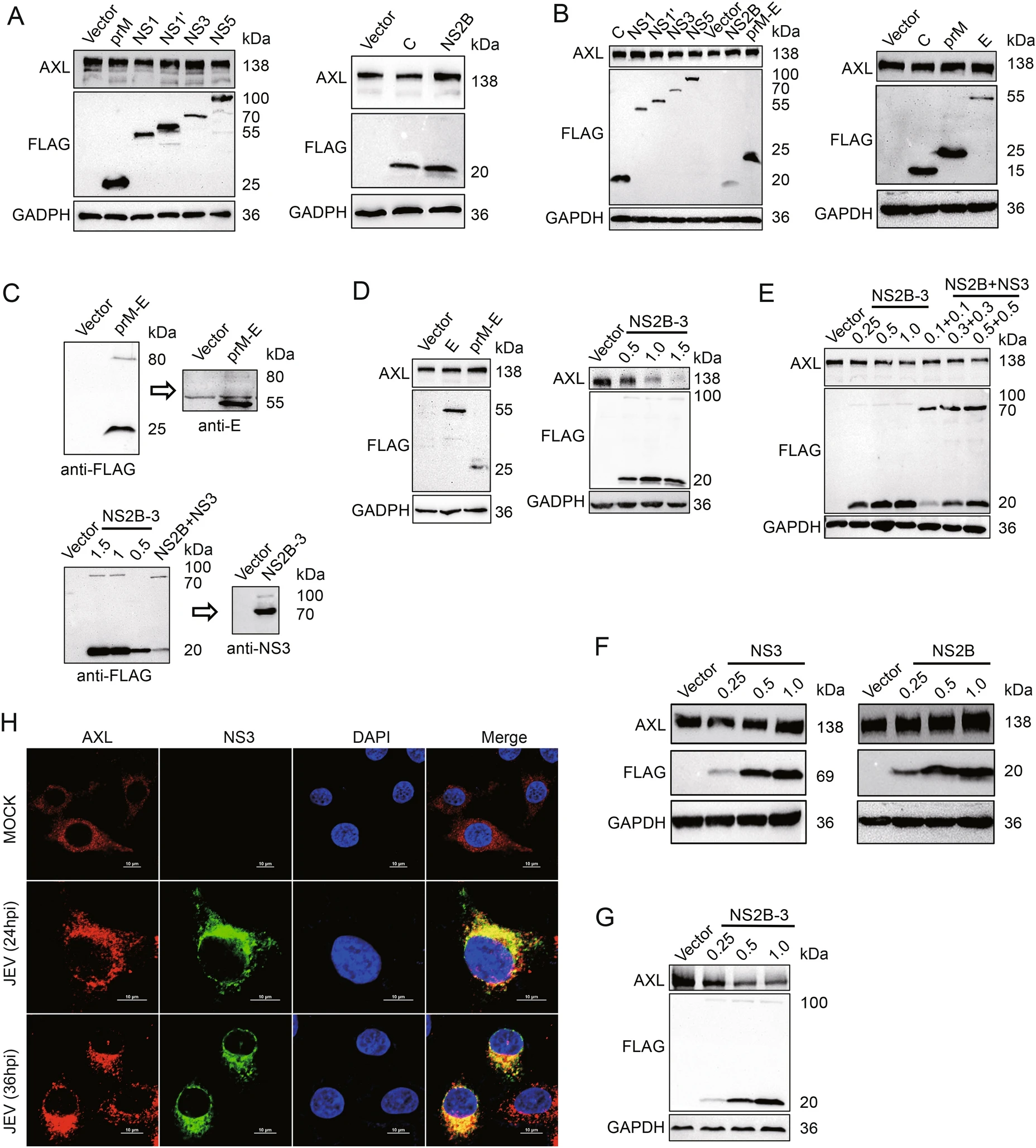
Japanese Encephalitis Virus NS2B-3 Protein Complex Promotes Cell Apoptosis and Viral Particle Release by Down-Regulating the Expression of AXL
2021, 36(6): 1503 doi: 10.1007/s12250-021-00442-3
Japanese encephalitis virus (JEV) is a flavivirus transmitted by mosquitoes that causes severe encephalitis in humans and animals. It has been suggested that AXL, a transmembrane protein, can promote the replication of various flaviviruses, such as dengue (DENV), Zika (ZIKV), and West Nile (WNV) viruses. However, the effect of AXL on JEV infection has not yet been determined. In the present study, we demonstrate that AXL is down-regulated after JEV infection in the late stage. JEV NS2B-3 protein specifically interacted with AXL, and promoted AXL degradation through the ubiquitin–proteasome pathway. AXL-degradation increased cell apoptosis by disrupting phosphatidylinositol 3-kinase (PI3K)/Akt signal transduction. In addition, the degradation of AXL promoted JEV release to supernatant, whereas the virus in the cell lysates decreased. The supplementation of AXL ligand Gas6 inhibited the JEV-mediated degradation of AXL. Altogether, we discover a new function of NS2B-3 during the process of JEV replication, and provide a new insight into the interactions between JEV and cell hosts.
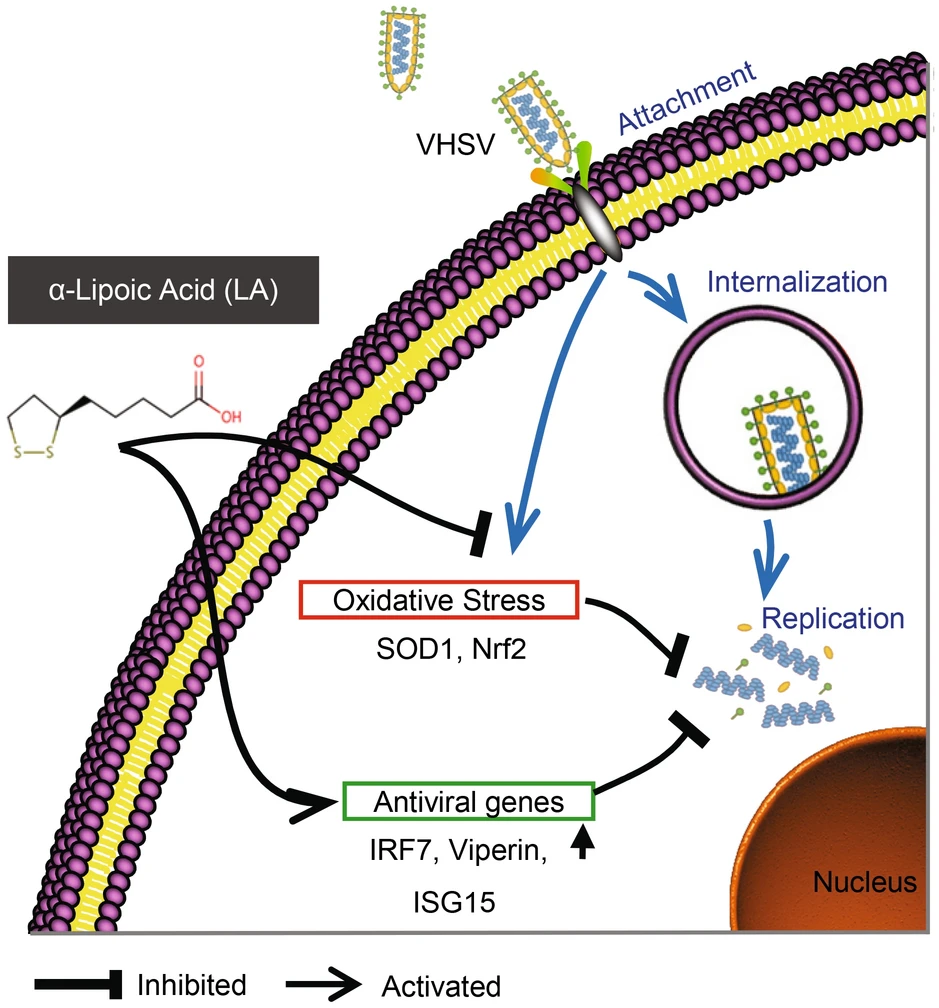
α-Lipoic Acid Exerts Its Antiviral Effect against Viral Hemorrhagic Septicemia Virus (VHSV) by Promoting Upregulation of Antiviral Genes and Suppressing VHSV-Induced Oxidative Stress
2021, 36(6): 1520 doi: 10.1007/s12250-021-00440-5
Viral hemorrhagic septicemia virus (VHSV), belonging to the genus Novirhabdovirus, Rhabdoviridae family, is a causative agent of high mortality in fish and has caused significant losses to the aquaculture industry. Currently, no effective vaccines, Food and Drug Administration-approved inhibitors, or other therapeutic intervention options are available against VHSV. α-Lipoic Acid (LA), a potent antioxidant, has been proposed to have antiviral effects against different viruses. In this study, LA (CC50 = 472.6 μmol/L) was repurposed to exhibit antiviral activity against VHSV. In fathead minnow cells, LA significantly increased the cell viability post-VHSV infection (EC50 = 42.7 μmol/L), and exerted a dose-dependent inhibitory effect on VHSV induced-plaque, cytopathic effects, and VHSV glycoprotein expression. The time-of-addition assay suggested that the antiviral activity of LA occurred at viral replication stage. Survival assay revealed that LA could significantly upregulated the survival rate of VHSV-infected largemouth bass in both co-injection (38.095% vs. 1.887%, P < 0.01) and post-injection manner (38.813% vs. 8.696%, P < 0.01) compared with the control group. Additional comparative transcriptome and qRT-PCR analysis revealed LA treatment upregulated the expression of several antiviral genes, such as IRF7, Viperin, and ISG15. Moreover, LA treatment reduced VHSV-induced reactive oxygen species production in addition to Nrf2 and SOD1 expression. Taken together, these data demonstrated that LA suppressed VHSV replication by inducing antiviral genes expression and reducing VHSV-induced oxidative stress. These results suggest a new direction in the development of potential antiviral candidate drugs against VHSV infection.
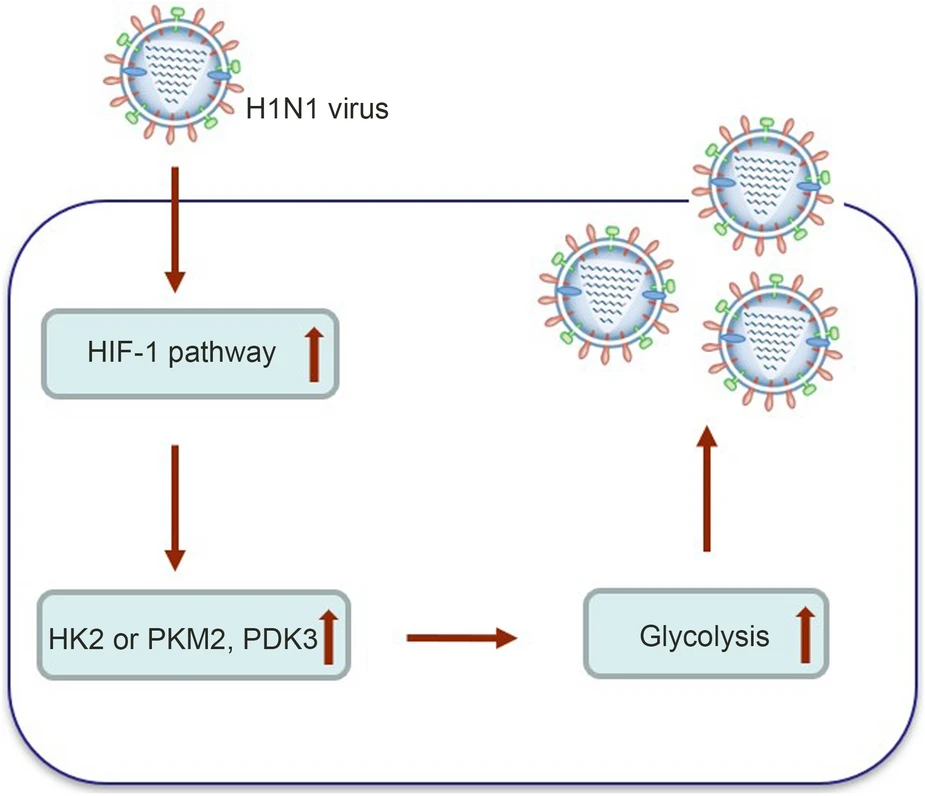
Influenza A Virus (H1N1) Infection Induces Glycolysis to Facilitate Viral Replication
2021, 36(6): 1532 doi: 10.1007/s12250-021-00433-4
Viruses depend on host cellular metabolism to provide the energy and biosynthetic building blocks required for their replication. In this study, we observed that influenza A virus (H1N1), a single-stranded, negative-sense RNA virus with an eight-segmented genome, enhanced glycolysis both in mouse lung tissues and in human lung epithelial (A549) cells. In detail, the expression of hexokinase 2 (HK2), the first enzyme in glycolysis, was upregulated in H1N1-infected A549 cells, and the expression of pyruvate kinase M2 (PKM2) and pyruvate dehydrogenase kinase 3 (PDK3) was upregulated in H1N1-infected mouse lung tissues. Pharmacologically inhibiting the glycolytic pathway or targeting hypoxia-inducible factor 1 (HIF-1), the central transcriptional factor critical for glycolysis, significantly reduced H1N1 replication, revealing a requirement for glycolysis during H1N1 infection. In addition, pharmacologically enhancing the glycolytic pathway further promoted H1N1 replication. Furthermore, the change of H1N1 replication upon glycolysis inhibition or enhancement was independent of interferon signaling. Taken together, these findings suggest that influenza A virus induces the glycolytic pathway and thus facilitates efficient viral replication. This study raises the possibility that metabolic inhibitors, such as those that target glycolysis, could be used to treat influenza A virus infection in the future.
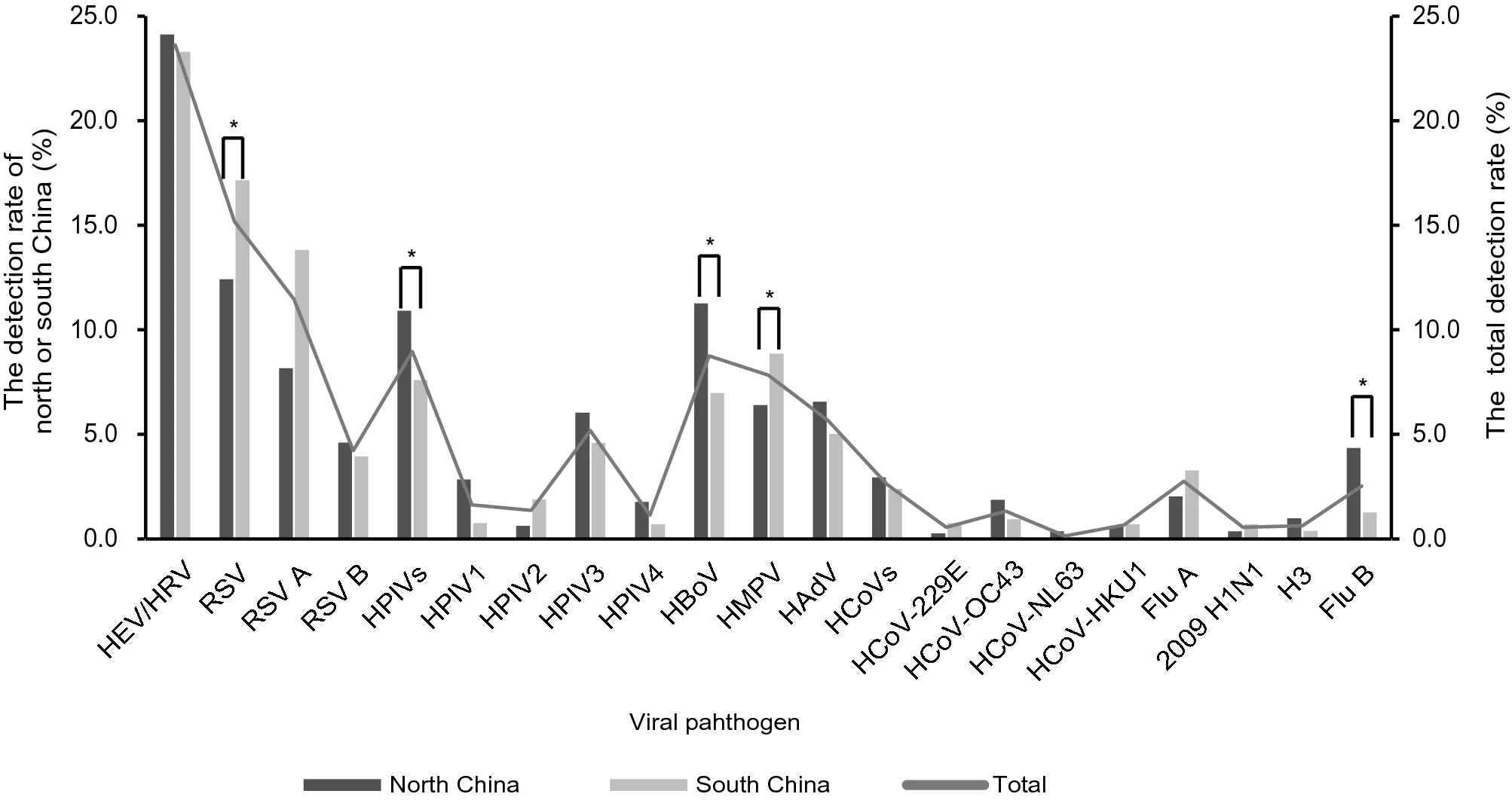
A Multicenter Study of Viral Aetiology of Community-Acquired Pneumonia in Hospitalized Children in Chinese Mainland
2021, 36(6): 1543 doi: 10.1007/s12250-021-00437-0
Community-acquired pneumonia (CAP) is one of the leading causes of morbidity and mortality in children worldwide. In this study, we aimed to describe the aetiology of viral infection of pediatric CAP in Chinese mainland. During November 2014 to June 2016, the prospective study was conducted in 13 hospitals. The hospitalized children under 18 years old who met the criteria for CAP were enrolled. The throat swabs or nasopharyngeal aspirates (NPAs) were collected which were then screened 18 respiratory viruses using multiplex PCR assay. Viral pathogens were present in 56.6% (1539/2721) of the enrolled cases, with the detection rate of single virus in 39.8% of the cases and multiple viruses in 16.8% of the cases. The most frequently detected virus was respiratory syncytial virus (RSV) (15.2%, 414/2721). The highest detection rate of virus was in\ 6-month-age group (70.7%, 292/413). RSV, human metapneumovirus (HMPV), human parainfluenza viruses (HPIVs) and influenza B virus (Flu B) showed the similar prevalence patterns both in north and south China, but HPIVs, Flu A, human bocavirus (HBoV), human adenovirus (HAdV) and human coronaviruses (HCoVs) showed the distinct circulating patterns in north and south China. Human enterovirus/human rhinovirus (HEV/HRV) (27.6%, 27/98), HBoV (18.4%, 18/98), RSV (16.3%, 16/98) and HMPV (14.3%, 14/98) were the most commonly detected viruses in severe pneumonia cases with single virus infection. In conclusion, viral pathogens are frequently detected in pediatric CAP cases and may therefore play a vital role in the aetiology of CAP. RSV was the most important virus in hospitalized children with CAP in Chinese mainland.
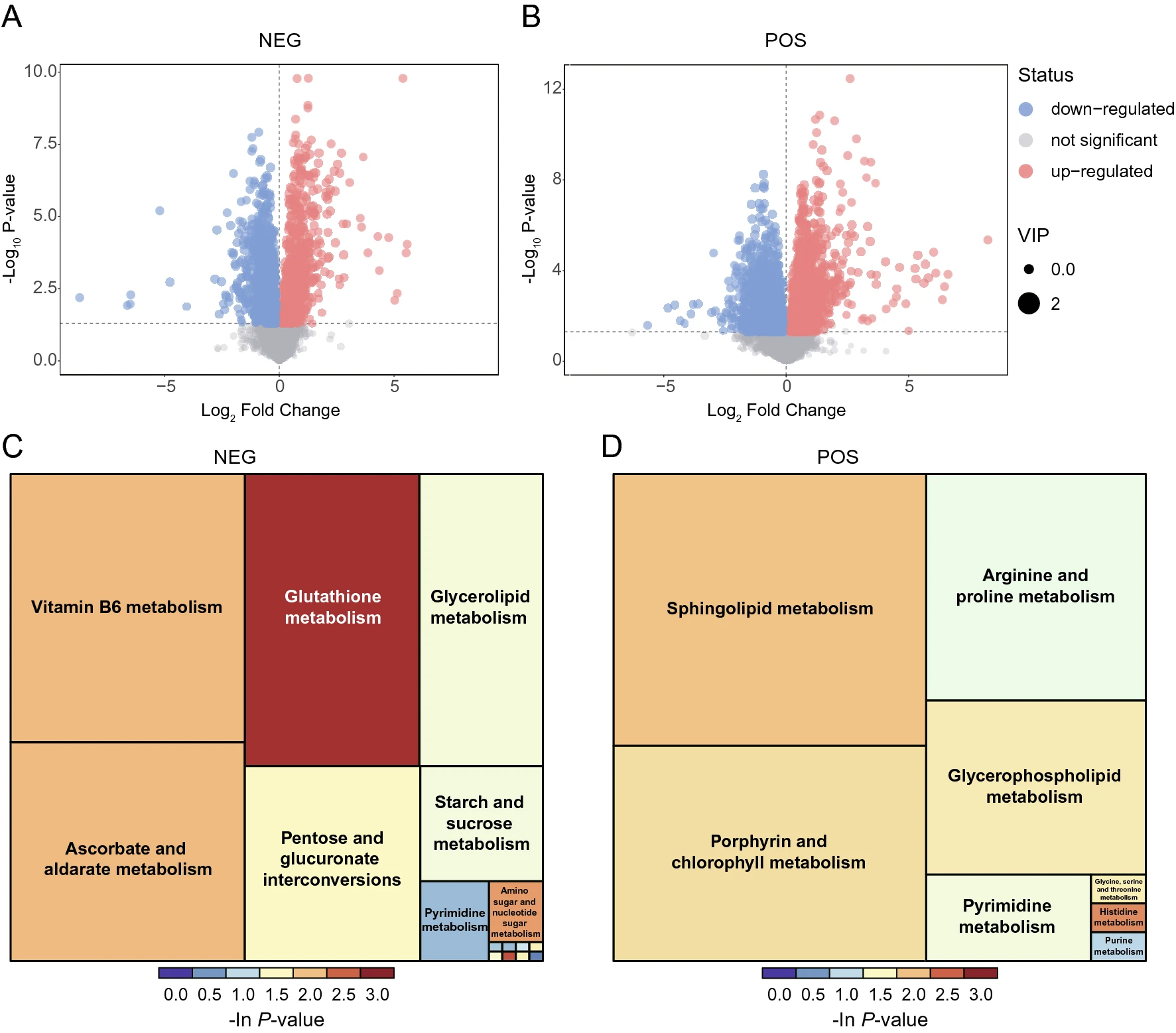
Integrated Metabolomics and Transcriptomics Analyses Reveal Metabolic Landscape in Neuronal Cells during JEV Infection
2021, 36(6): 1554 doi: 10.1007/s12250-021-00445-0
Japanese encephalitis virus (JEV) is a leading cause of viral encephalitis in endemic regions of Asia. The neurotropism of JEV and its high-efficiency replication in neurons are the key events for pathogenesis. Revealing the interplay between virus and host cells in metabolic facet is of great importance both for unraveling the pathogenesis mechanisms and providing novel antiviral targets. This study took advantage of the integration analysis of metabolomics and transcriptomics to depict the metabolic profiles of neurons during the early stage of JEV infection. Increased glycolysis and its branched pentose phosphate pathway (PPP) flux and impaired oxidative phosphorylation (OXPHOS) in glucose utilization, and the catabolic patterns of lipid metabolism were created to facilitate the biosynthesis of precursors needed for JEV replication in neurons. Pharmacological inhibitions of both glycolysis pathway and PPP in neurons suggested its indispensable role in maintaining the optimal propagation of JEV. In addition, analysis of metabolomic-transcriptomic regulatory network showed the pivotal biological function of lipid metabolism during JEV infection. Several pro-inflammatory lipid metabolites were significantly up-regulated and might partially be responsible for the progression of encephalitis. These unique metabolic reprogramming features might give deeper insight into JEV infected neurons and provide promising antiviral approaches targeting metabolism.
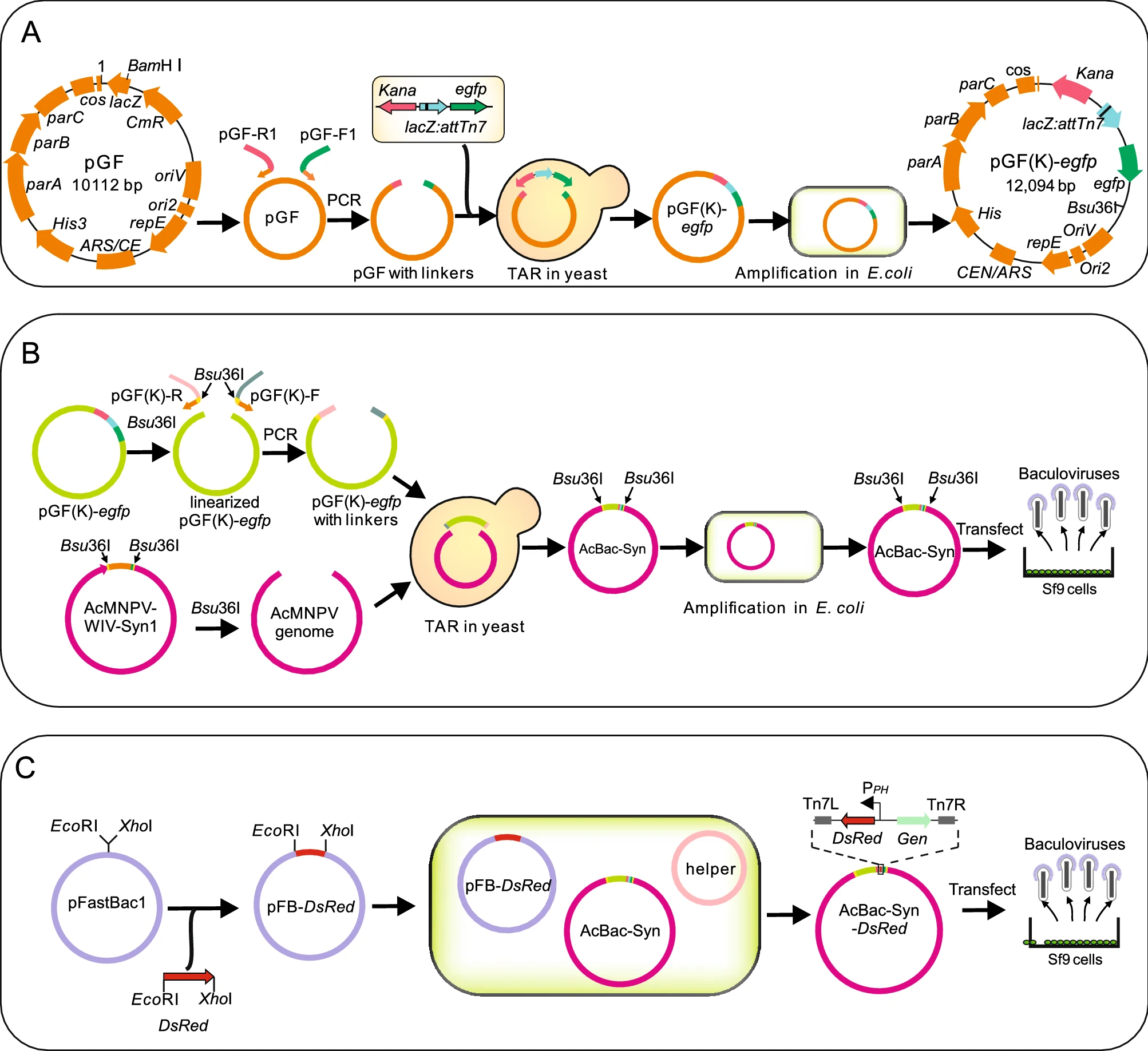
Construction and Characterization of a Novel Bacmid AcBac-Syn Based on a Synthesized Baculovirus Genome
2021, 36(6): 1566 doi: 10.1007/s12250-021-00449-w
Baculoviruses are large DNA viruses which have been widely used as expression vectors and biological insecticides. Homologous recombination and Bac-to-Bac system have been the main methods for manipulating the baculovirus genome. Recently, we generated a synthetic baculovirus AcMNPV-WIV-Syn1 which fully resembled its parental virus Autographa californica multiple nucleopolyhedrovirus (AcMNPV). Here, we report the modification of AcMNPV-WIV-Syn1 into a novel bacmid, AcBac-Syn, which can be used as a backbone for Bac-to-Bac system. To achieve this, a vector contained a LacZ:attTn7 and egfp cassette was constructed, and recombined with a linearized AcMNPV-WIV-Syn1 genome by transformation-associated recombination in yeast to generate bacmid AcBac-Syn. The bacmid was then transfected to insect cells and the rescued virus showed similar biological characteristics to the wild-type virus in terms of the kinetics of budded virus production, the morphology of occlusion bodies, and the oral infectivity in insect larvae. For demonstration, a red fluorescent protein gene Dsred was transposed into the attTn7 site by conventional Bac-to-Bac method, and the transfection and infection assays showed that AcBac-Syn can be readily used for foreign gene insertion and expression. AcBac-Syn has several advantages over the conventional AcMNPV bacmids, such as it contains an egfp reporter gene which facilitates visualization of virus propagation and titration; its DNA copy numbers could be induced to a higher level in E. coli; and the retaining of the native polyhedrin gene in the genome making it an attractive system for studying the functions of gene related to occlusion body assembly and oral infection.
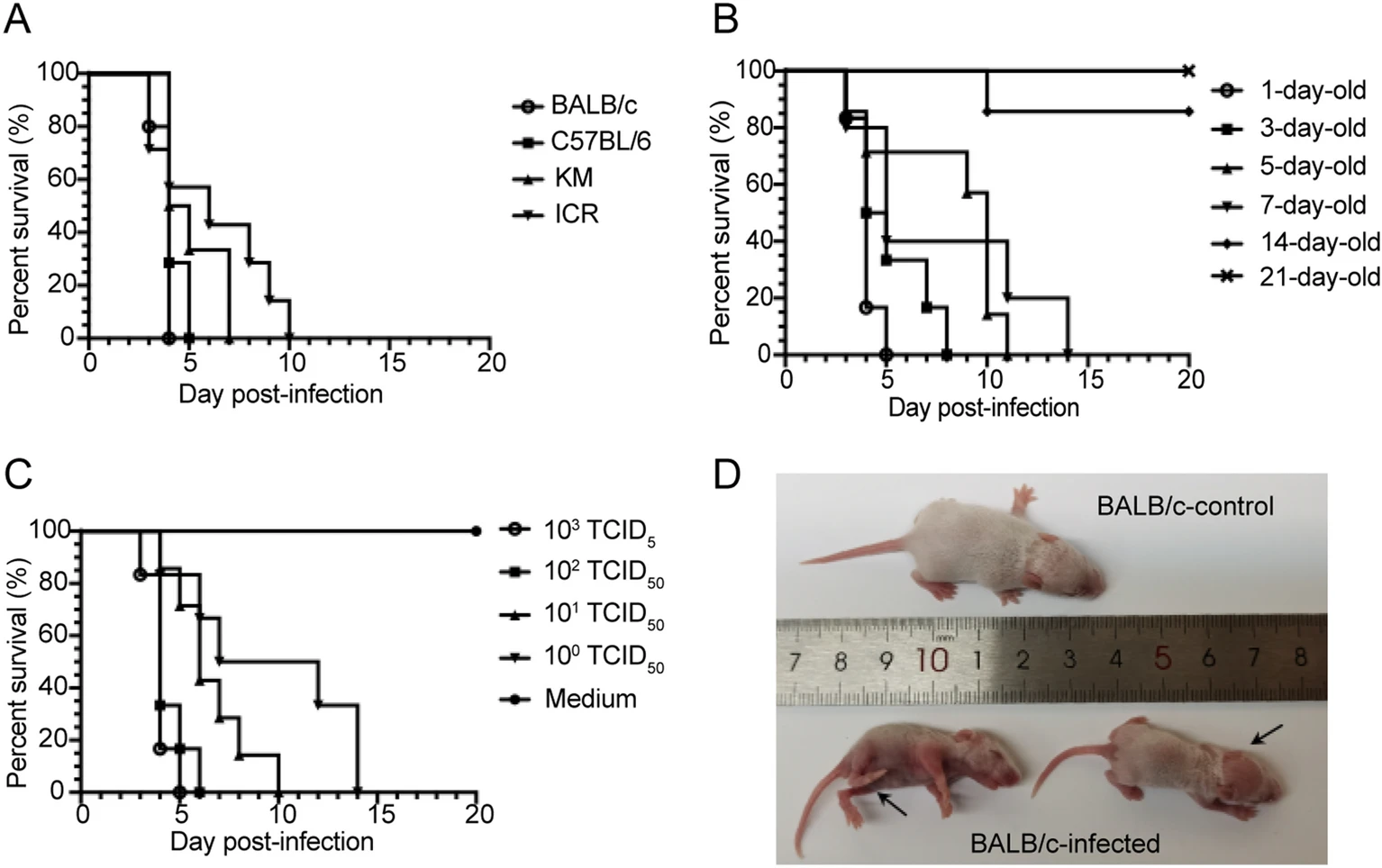
Development of A Neonatal Mouse Model for Coxsackievirus B1 Antiviral Evaluation
2021, 36(6): 1575 doi: 10.1007/s12250-021-00444-1
Coxsackievirus B1 (CVB1) is a leading causative agent of severe infectious diseases in humans and has been reported to be associated with outbreaks of aseptic meningitis, myocarditis, and the development of chronic diseases such as type 1 diabetes mellitus (T1DM). There is no approved vaccine or effective antiviral therapy to treat CBV1 infection. And animal models to assess the effects of antiviral agents and vaccine remain limited. In this study, we established a neonatal mouse model of CVB1 using a clinically isolated strain to characterize the pathological manifestations of virus infection and to promote the development of vaccines and antiviral drugs against CVB1. One-day-old BALB/c mice were susceptible to CVB1 infection by intraperitoneal injection. Mice challenged with CVB1 at a low dose [10 median tissue culture infective dose (TCID50)] exhibited a series of clinical symptoms, such as inactivity, emaciation, limb weakness, hair thinning, hunching and even death. Pathological examination and tissue viral load analysis showed that positive signals of CVB1 were detected in the heart, spinal cord, limb muscle and kidney without pathological damage. Particularly, CVB1 had a strong tropism towards the pancreas, causing severe cellular necrosis with inflammatory infiltration, and was spread by viraemia. Notably, the monoclonal antibody (mAb) 6H5 and antisera elicited from CVB1-vaccinated mice effectively protected the mice from CVB1 infection in the mouse model. In summary, the established neonatal mouse model is an effective tool for evaluating the efficacy of CVB1 antiviral reagents and vaccines.

MicroRNA-324-3p Plays A Protective Role Against Coxsackievirus B3-Induced Viral Myocarditis
2021, 36(6): 1585 doi: 10.1007/s12250-021-00441-4
Viral myocarditis (VM) is an inflammatory disease of the myocardium associated with heart failure, which is caused by common viral infections. A majority of the infections are initiated by coxsackievirus B3 (CVB3). MicroRNAs (miRNAs) have a major role in various biological processes, including gene expression, cell growth, proliferation, and apoptosis, as well as viral infection and antiviral immune responses. Although, miRNAs have been found to regulate viral infections, their role in CVB3 infection remains poorly understood. In the previous study, miRNA microarray results showed that miR-324-3p expression levels were significantly increased when cells and mice were infected with CVB3. It was also found that miR-324-3p downregulated TRIM27 and decreased CVB3 replication in vitro and in vivo. In vitro, analysis of downstream signaling of TRIM27 revealed that, miR-324-3p inhibited CVB3 infection, and reduced cytopathic effect and viral plaque formation by reducing the expression of TRIM27. In vivo, miR-324-3p decreased the expression of TRIM27, reduced cardiac viral replication and load, thereby strongly attenuating cardiac injury and inflammation. Taken together, this study suggests that miR-324-3p targets TRIM27 to inhibit CVB3 replication and viral load, thereby reducing the cardiac injury associated with VM.
Identification and Characterization of a Novel Single Domain Antibody Against Ebola Virus
2021, 36(6): 1600 doi: 10.1007/s12250-021-00454-z
Ebola virus (EBOV) belongs to the Filoviridae family and causes severe illnesses such as hemorrhagic fever with a high mortality rate up to 90%. Now two antibody drugs termed Inmazeb and Ebanga have been approved for treating EBOV infection. However, clinical studies have demonstrated that the mortality rate of the patients who received these two antibody drugs remains above 30%. Therefore, novel therapeutics with better efficacy is still desired. The isolated human IgG1 constant domain 2 (CH2 domain) has been proposed as a scaffold for the development of C-based single domain antibodies (C-sdAbs) as therapeutic candidates against viral infections and other diseases. Here, we screened and identified a novel C-sdAb termed M24 that targets EBOV glycoprotein (GP) from a C-sdAb phage display library. M24 neutralizes the pseudotype EBOV with IC50 of 0.8 nmol/L (12 ng/mL) and has modest neutralizing activity against authentic EBOV. Epitope determination, including molecular docking and site mutation analysis, discloses that M24 binds to the internal fusion loop (IFL) within GP2, a transmembrane subunit of GP. Interestingly, we found that the binding of M24 to GP at pH 5.5 has dramatically decreased compared to the binding at pH 7.5, which may lead to weak efficacy in the neutralization of authentic EBOV. Since no sdAb against EBOV infection has been reported to date, our results not only give a proof of concept that sdAbs could be utilized for the development of potential therapeutic candidates against EBOV infection, but also provide useful information for the discovery and improvement of anti-EBOV agents.
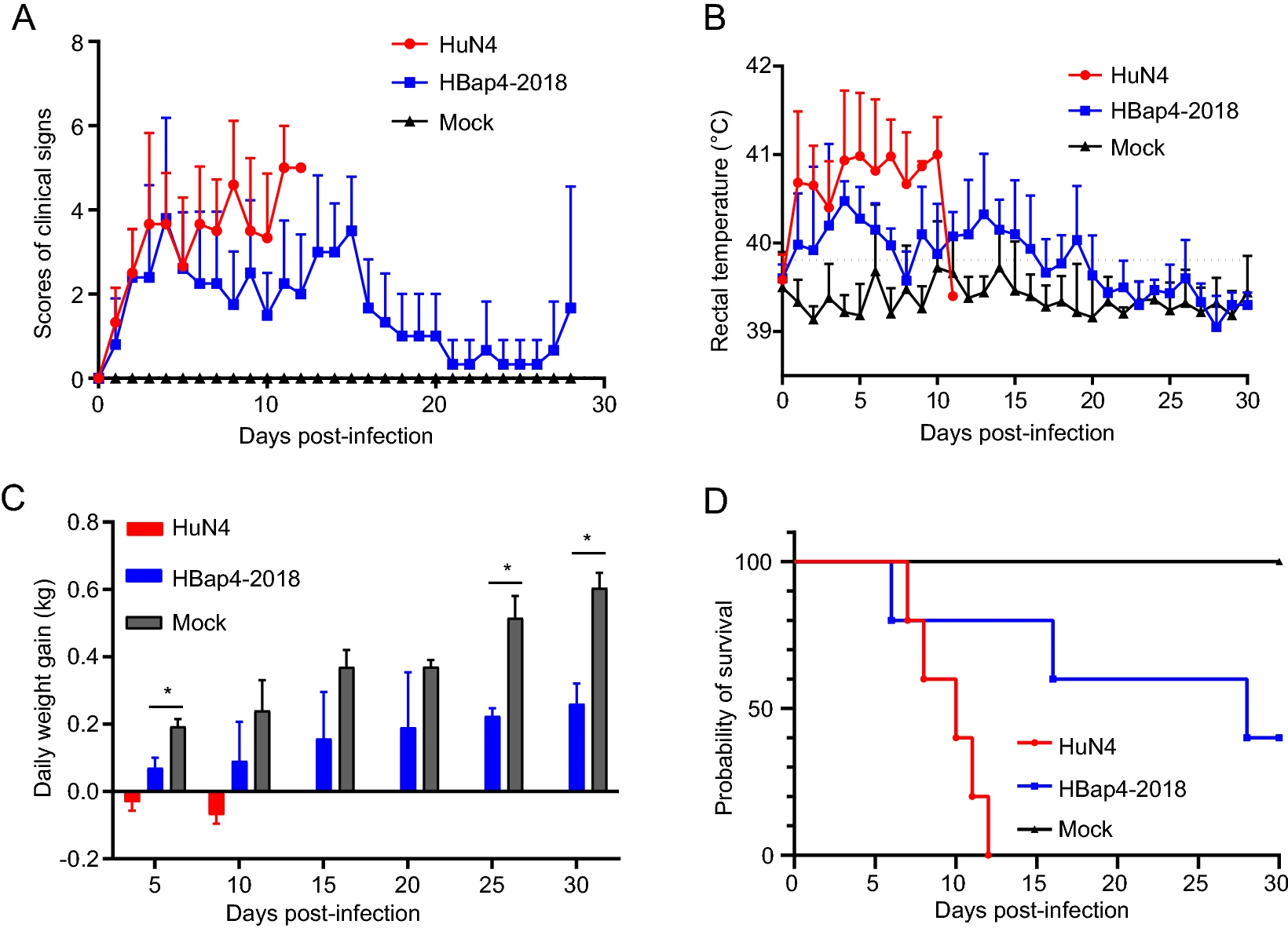
The Novel PRRSV Strain HBap4-2018 with a Unique Recombinant Pattern Is Highly Pathogenic to Piglets
2021, 36(6): 1611 doi: 10.1007/s12250-021-00453-0
Currently, various porcine reproductive and respiratory syndrome virus (PRRSV) variants emerged worldwide with different genetic characteristics and pathogenicity, increasing the difficulty of PRRS control. In this study, a PRRSV strain named HBap4-2018 was isolated from swine herds suffering severe respiratory disease with high morbidity in Hebei Province of China in 2018. The genome of HBap4-2018 is 15,003 nucleotides in length, and compared with NADC30-like PRRSV, nsp2 of HBap4-2018 has an additional continuous deletion of five amino acids. Phylogenetic analysis based on complete genome and ORF5 showed that HBap4-2018 belonged to lineage 8 of PRRSV-2, which was characterized by highly variable genome. However, HBap4-2018 was classified into lineage 1 based on phylogenetic analysis of nsp2, sharing higher amino acid homology (85.3%–85.5%) with NADC30-like PRRSV. Further analysis suggested that HBap4-2018 was a novel natural recombinant PRRSV with three recombinant fragments in the genome, of which highly pathogenic PRRSV (HP-PRRSV) served as the major parental strains, while NADC30-like PRRSV served as the minor parental strains. Five recombination break points were identified in nsp2,nsp3,nsp5,nsp9 and ORF6, respectively, presenting a novel recombinant pattern in the genome. Piglets inoculated with HBap4-2018 presented typical clinical signs with a mortality rate of 60%. High levels of viremia and obvious macroscopic and histopathological lesions in the lungs were observed, revealing the high pathogenicity of HBap4-2018 in piglets.
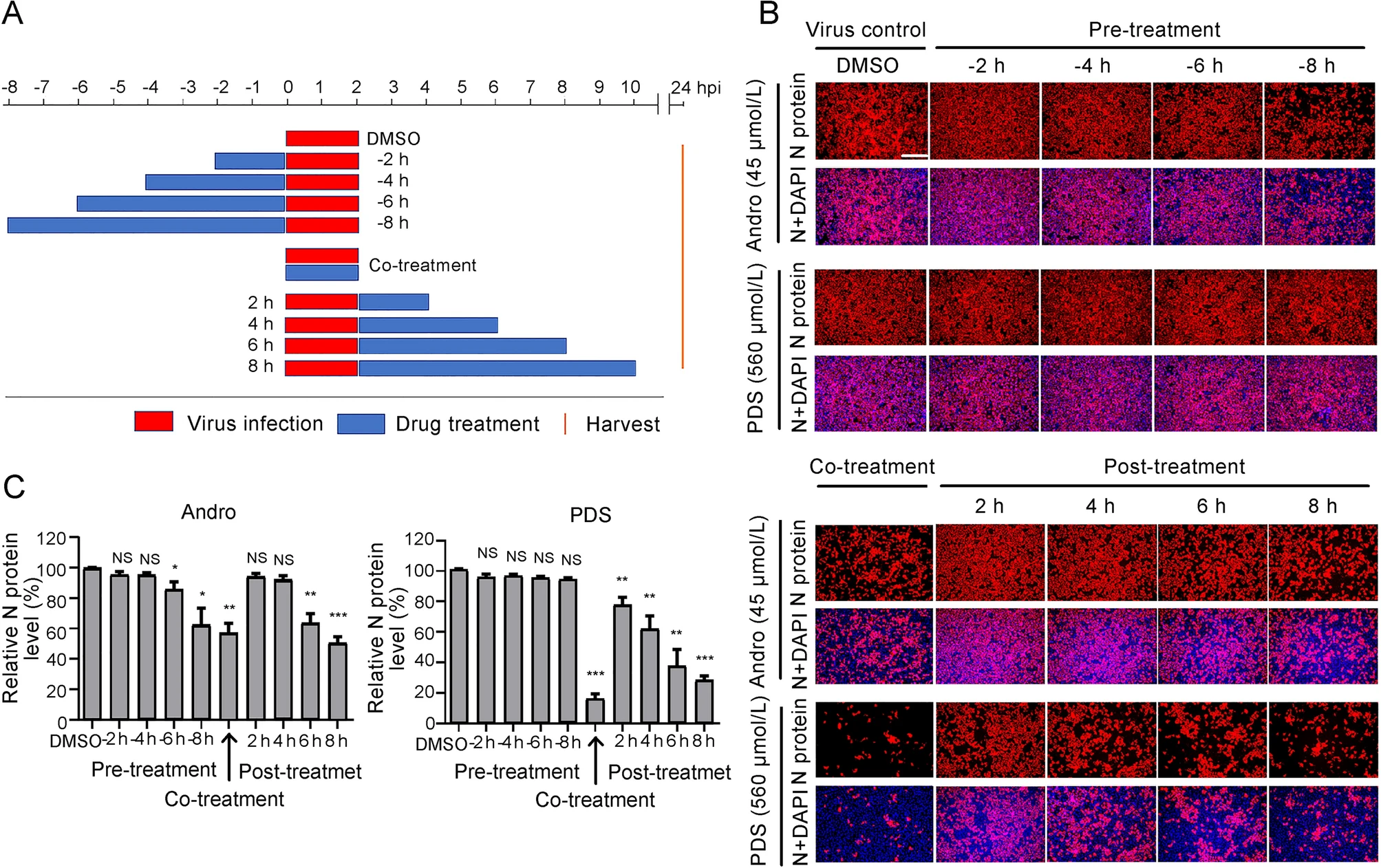
Andrographolide and Its Derivative Potassium Dehydrographolide Succinate Suppress PRRSV Replication in Primary and Established Cells via Differential Mechanisms of Action
2021, 36(6): 1626 doi: 10.1007/s12250-021-00455-y
Porcine reproductive and respiratory syndrome virus (PRRSV) continues to cause significant economic loss worldwide and remains a serious threat to the pork industry. Currently, vaccination strategies provide limited protection against PRRSV infection, and consequently, new antiviral strategies are urgently required. Andrographolide (Andro) and its derivative potassium dehydrographolide succinate (PDS) have been used clinically in China and other Asian countries as therapies for inflammation-related diseases, including bacterial and viral infections, for decades. Here, we demonstrate that Andro and PDS exhibit robust activity against PRRSV replication in Marc-145 cells and primary porcine alveolar macrophages (PAMs). The two compounds exhibited broad-spectrum inhibitory activities in vitro against clinically circulating type 2 PRRSV GD-HD, XH-GD, and NADC30-like HNhx strains in China. The EC50 values of Andro against three tested PRRSV strain infections in Marc-145 cells ranged from 11.7 to 15.3 μmol/L, with selectivity indexes ranging from 8.3 to 10.8, while the EC50 values of PDS ranged from 57.1 to 85.4 μmol/L, with selectivity indexes ranging from 344 to 515. Mechanistically, the anti-PRRSV activity of the two compounds is closely associated with their potent suppression on NF-κB activation and enhanced oxidative stress induced by PRRSV infection. Further mechanistic investigations revealed that PDS, but not Andro, is able to directly interact with PRRSV particles. Taken together, our findings suggest that Andro and PDS are promising PRRSV inhibitors in vitro and deserves further in vivo studies in swine.

Development and Characterization of SYBR Green Ⅰ Based RT-PCR Assay for Detection of Omsk Hemorrhagic Fever Virus
2021, 36(6): 1644 doi: 10.1007/s12250-021-00389-5
The Application of a Safe Neutralization Assay for Ebola Virus Using Lentivirus-Based Pseudotyped Virus
2021, 36(6): 1648 doi: 10.1007/s12250-021-00405-8

New Simian Enterovirus 19 (EV-A122) Strains in China Reveal Large-Scale Inter-Serotype Recombination between Simian EV-As
2021, 36(6): 1652 doi: 10.1007/s12250-021-00412-9

Genomic Characterization of a New Coronavirus from Migratory Birds in Jiangxi Province of China
2021, 36(6): 1656 doi: 10.1007/s12250-021-00402-x
Genetic Variations in Human Parechovirus Type 3 in Infants with Central Nervous System Infection
2021, 36(6): 1660 doi: 10.1007/s12250-021-00426-3

Inhibition of the Neddylation Pathway Suppresses Enterovirus Replication
2021, 36(6): 1664 doi: 10.1007/s12250-021-00427-2

The Establishment and Spatiotemporal History of A Novel HIV-1 CRF01_AE Lineage in Shenyang City, Northeastern China in 2002–2019
2021, 36(6): 1668 doi: 10.1007/s12250-021-00435-2
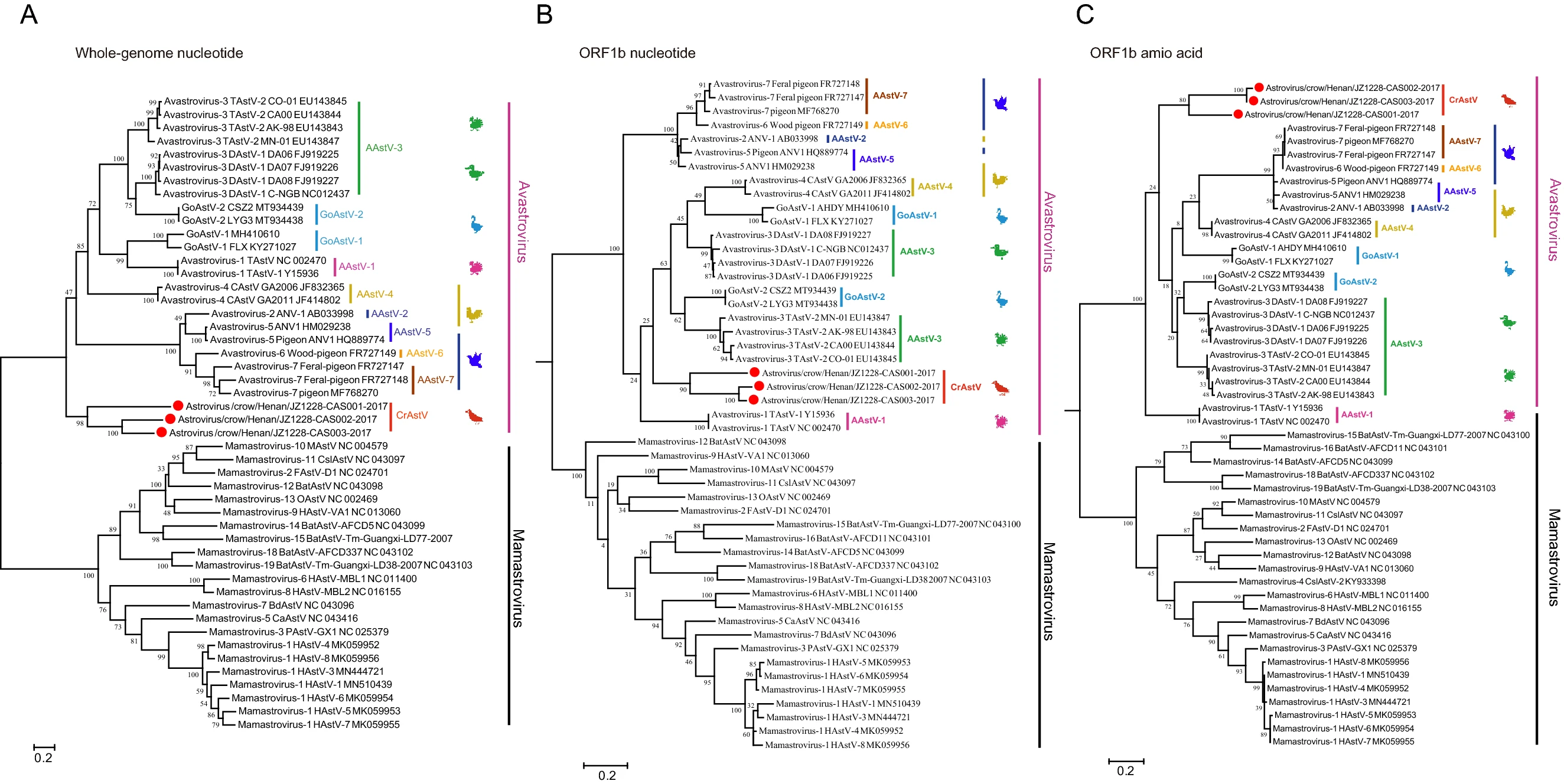
Three Novel Avastroviruses Identified in Dead Wild Crows
2021, 36(6): 1673 doi: 10.1007/s12250-021-00416-5

Construction of an Infectious Clone for Mosquito-Derived Tembusu Virus Prototypical Strain
2021, 36(6): 1678 doi: 10.1007/s12250-021-00447-y
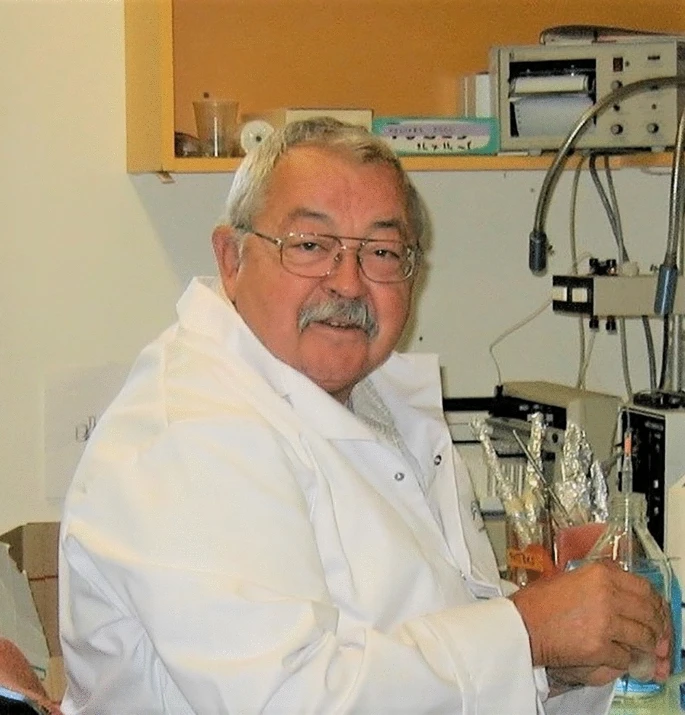
In Memory of Dr. Jean-Robert Bonami
2021, 36(6): 1682 doi: 10.1007/s12250-021-00456-x
Correction to: Establishment of a Reverse Genetic System of Severe Fever with Thrombocytopenia Syndrome Virus Based on a C4 Strain
2021, 36(6): 1683 doi: 10.1007/s12250-021-00420-9

Correction to: In Memory of Dr. Jean-Robert Bonami
2021, 36(6): 1685 doi: 10.1007/s12250-021-00459-8
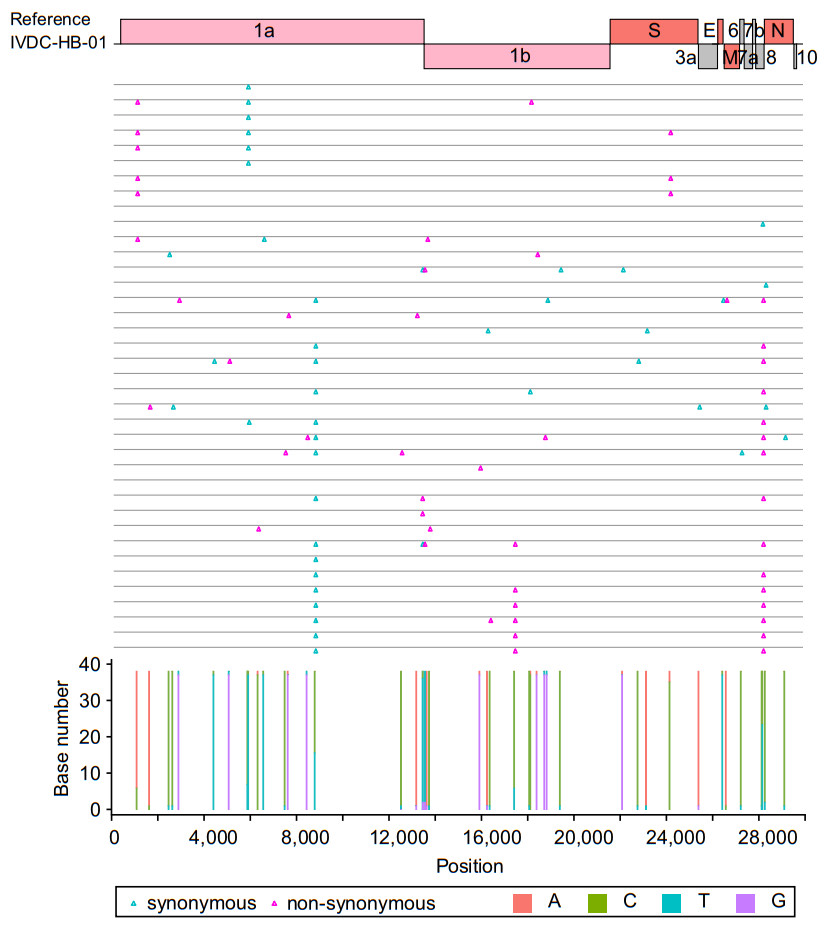
Correction to: Rapid Acquisition of High-Quality SARS-CoV-2 Genome via Amplicon-Oxford Nanopore Sequencing
2021, 36(6): 1686 doi: 10.1007/s12250-021-00465-w
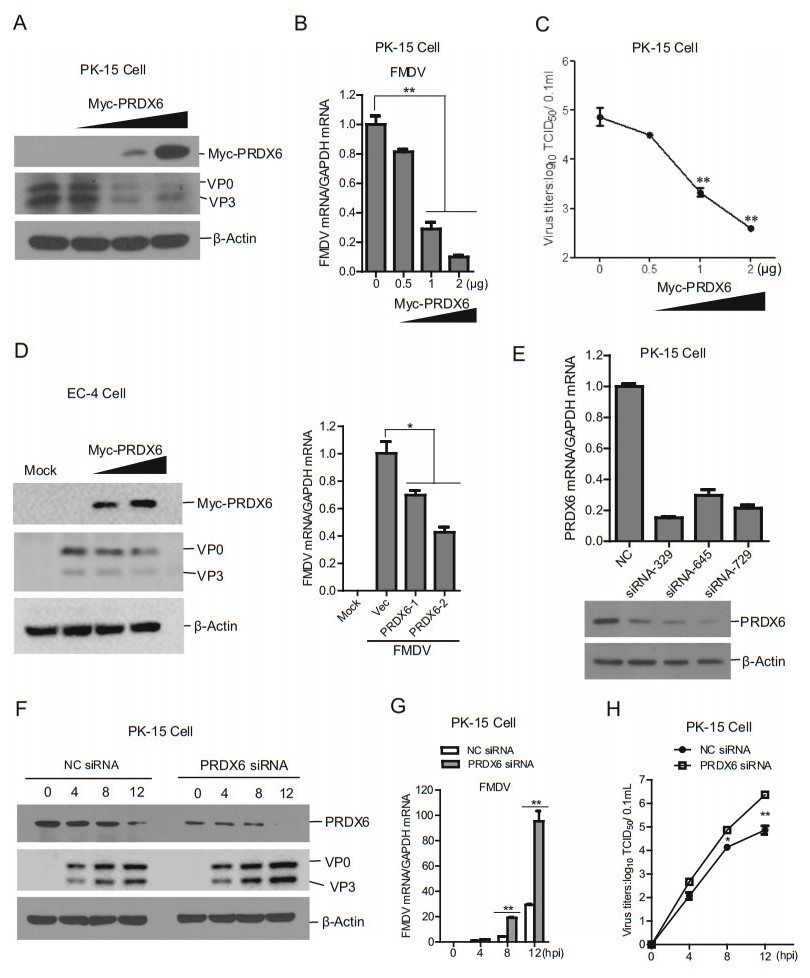
Correction to: Porcine Picornavirus 3C Protease Degrades PRDX6 to Impair PRDX6-mediated Antiviral Function
2021, 36(6): 1688 doi: 10.1007/s12250-021-00466-9
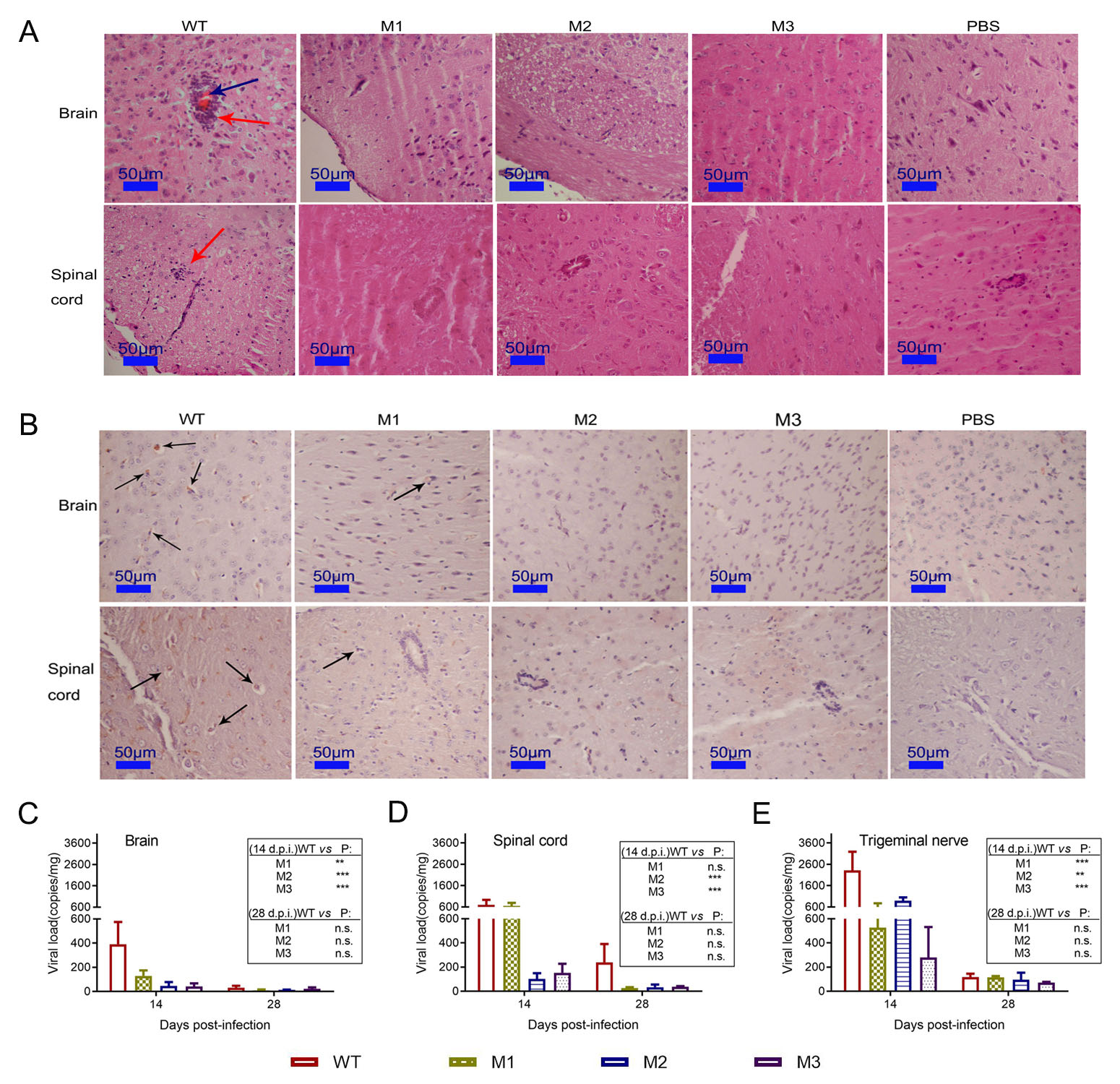
Correction to: Attenuated phenotypes and analysis of a herpes simplex virus 1 strain with partial deletion of the UL7, UL41 and LAT genes
2021, 36(6): 1690 doi: 10.1007/s12250-021-00467-8
Correction to: Replication of LZTFL1 Gene Region as a Susceptibility Locus for COVID-19 in Latvian Population
2021, 36(6): 1692 doi: 10.1007/s12250-021-00464-x

Correction to: Integrated Metabolomics and Transcriptomics Analyses Reveal Metabolic Landscape in Neuronal Cells During JEV Infection
2021, 36(6): 1693 doi: 10.1007/s12250-021-00468-7