-
Correction to:
Virologica Sinica (2021) 36:901–912
https://doi.org/10.1007/s12250-021-00378-8
The original version of article contains errors in Figure 4, Supplementary Figure S2, Supplementary Table S2, and the corresponding description in part 3 of "Results" section. The corrected contents are shown below:
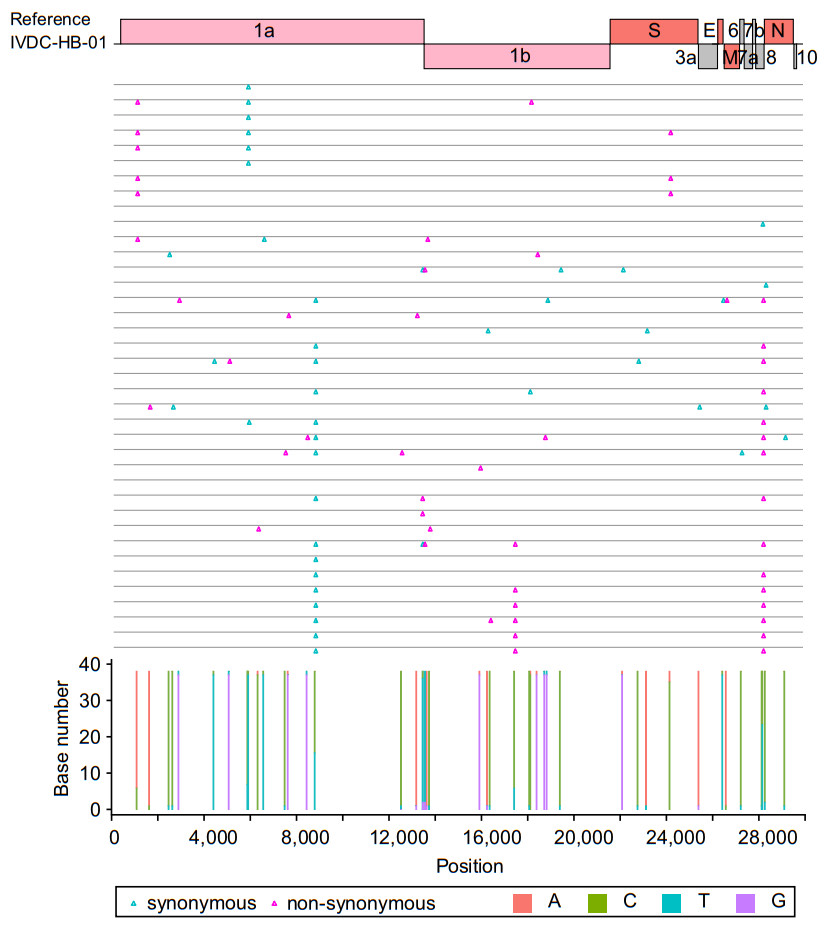
Figure 4. Nucleotide and amino acid variations in 38 sample genomes. The top shows the open reading frames (ORFs) position of the reference genome, and the middle displays the amino acid variations on the genome of each sample. The blue triangle represents synonymous mutation, the red triangle represents non-synonymous mutation. The bottom shows the nucleotide composition at the mutation sites identified in the38 samples. Different colors represent different bases, the bar in red is adenine (A), the bar in green is cytosine (C), the bar in blue is thymine (T), and the bar in purple is guanine (G)
1. Figure 4
2. Supplementary Figure S2
3. Supplementary Table S2
4. Part 3 of the "Results" section:
Subtitle: High-quality Genome Revealed Genetic Variations of SARS-CoV-2
By using bcftools mpileup and Medaka for SNP calling and a published script (margin_cons.py; Quick et al. 2017) for consensus generation, 38 nearly full-length (on average about 99.61%) SARS-CoV-2 genomes with high-quality, and 4 shorter genomes (from 93.92 to 97.68%) with gaps were obtained. All these genomes have been deposited in NGDC (accessions: GWHALPE01000000-GWHALPT010 00000 and GWHALRI01000000-GWHALSH01000000) and GISAID database (accessions: EPI_ISL_493149- EPI_ISL_493190). Compared to an early virus isolate genome IVDC-HB-01 (GISAID accession number: EPI_ISL_402119), 44 SNP sites were discovered in the 38 nearly full-length genomes, with 21 synonymous mutation sites and 23 nonsynonymous ones (Fig. 4, Supplementary Table S2). For each virus genome, there were 0 to 5 SNPs compared to IVDC-HB-01. For all SNP sites, only one mutated nucleotide existed. Through comparison with all deposited genome sequences in GISAID database, as of submitted date of March 1, 2020, 7 of the SNPs were appeared in previous released genome sequences (before the date of the given sample collected) as well as observed in the following virus genomes, 37 SNPs were firstly discovered at the date of sample collection (Supplementary Fig. S2, Supplementary Table S2).
The original article can be found online at https://doi.org/10.1007/s12250-021-00378-8.
Correction to: Rapid Acquisition of High-Quality SARS-CoV-2 Genome via Amplicon-Oxford Nanopore Sequencing
- Published Date: 17 November 2021
Abstract:














 DownLoad:
DownLoad: