HTML
-
The yellow fever virus (YFV) belongs to the genus Fla-vivirus of the family Flaviviridae. Mainly transmitted by the bite of Aedes mosquitoes, YFV can cause devastating yellow fever disease, which was once a great threat to public health in the history. Although the live attenuated vaccine 17D has been used for over eighty years (Norrby 2007; Barrett 2017), it has been estimated that there are still tens of thousands of deaths caused by YF annually in the tropical area (Garske et al. 2014; WHO 2019). Recent outbreaks in Africa and South America implied the re-emerging of YF (Goldani 2017; Zhao et al. 2018). Notably, the risk of YFV global expanding has increased in recent years (Cui et al. 2017).
Although YFV is the prototype member of the genus Flavivirus, far less is known about its replication mecha-nisms when compared with its cousins, such as dengue virus and Zika virus (Gebhard et al. 2011; Göertz et al. 2017; Liu and Qin 2020). Furthermore, there is no clinically-approved anti-viral medicine against flaviviruses, including YFV. Thus, there is an urgent need of novel tools to study YFV replication and to develop anti-YFV drugs. To address this issue, yellow fever reporter virus, which provides a rapid and convenient way to monitor viral propagation should be developed.
The genome of YFV is a single-stranded, positive-sense RNA molecule about 11 kb (kilobase) in length. A single open reading frame (ORF) in viral genome encodes a polyprotein precursor with over 3, 400 residues, which is further processed into three structural proteins (capsid protein C, pre-membrane/membrane protein prM/M and envelope protein E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). The 5' and 3' untranslated regions (UTRs) of YFV are highly structured, and contain numerous proposed cis-acting RNA elements (CREs). As inferred from the findings in other flaviviruses (Chapman et al. 2014; Villordo et al. 2015; Liu et al. 2016; Xie et al. 2019), The YFV CREs are supposed to play critical roles during viral RNA synthesis and other stages in virus life-cycle, but only a few CREs of YFV had been experimentally characterized (Bredenbeek et al. 2003; Silva et al. 2010).
In this work, we introduced the luciferase gene from Renilla reniformis (Rluc) into an infectious clone of YFV 17D strain to generate the 17D-Rluc.2A reporter virus. The 17D-Rluc.2A expressed high levels of luciferase in both mammalian and mosquito cells, and a good correlation between luciferase expression and progeny virus produc-tion was demonstrated. Mutagenesis analysis of the con-served 5' -SLA element demonstrated that the 17D-Rluc.2A is of great value for the investigation of YFV replication. Furthermore, we showed that the luciferase signals in 17D-Rluc.2A-infected cells were highly sensitive and specific to treatment of NITD008 (Yin et al. 2009), an inhibitor of flavivirus NS5 RNA-dependent RNA polymerase (RdRp). We then provided evidence that nanchangmycin (Rausch et al. 2017) can efficiently inhibit YFV infection in vitro using the 17D-Rluc.2A reporter virus. Our findings demonstrated that 17D-Rluc.2A is a powerful tool for investigation of YFV replication and evaluation of antivirals.
-
BHK-21 cells (ATCC CCL-10) were cultured in Dul-becco's modified Eagle's medium (DMEM) supplemented with 5% Fetal Bovine Serum (FBS, Biowest), 10 mmol/L HEPES and 1% Penicillin–streptomycin (PS). A549 (ATCC CCL-185) and C6/36 cells (ATCC CRT- 1660) were cultured in Roswell Park Memorial Institute 1640 medium (RPMI 1640) supplemented with 10% FBS, 25 mmol/L HEPES and 1% PS. BHK-21 and A549 cells were cultured in a 37 ℃ incubator with 5% CO2. C6/36 cells were cultured at 28 ℃.
17D and 17D-Rluc.2A were propagated in BHK-21 cells. After the appearance of visible cytopathic effect (CPE), the supernatants were collected with centrifugation at 3000 ×g for 10 min to pellet cellular debris and then stored as single-use aliquots at - 80 ℃. The virus titer was determined by plaque assay as described before (Yuan et al. 2017).
-
DNA manipulations were performed by standard molecular cloning technologies. The Not I-SP6 promoter-17D 5'- UTR-C117 fragment was amplified using the pACNR-FLYF17Dx (Bredenbeek et al. 2003) as the template. Then, the coding sequences of Rluc and foot and mouth disease virus (FMDV) 2A peptide, which is followed by the re-coded coding region for the N-terminal 18 residues of the 17D C protein was amplified from the pACNR-17D-Rluc-Rep plasmid (manuscript in preparation). These two frag-ments were assembled to generate the Not I-Rluc.2A-SnaB I fragment by overlapping PCR method. Then the AT-cloned Not I-Rluc.2A-SnaB I fragment was inserted into the pACNR-Nluc-YF17D plasmid (manuscript in prepa-ration) by Not I/SnaB I sub-cloning to obtain the pACNR-17D-Rluc.2A-ic plasmid. The 5' -SLA mutants were gen-erated by similar strategies. The pACNR-17D-Rluc.2A-ic-ΔGDD plasmid was constructed by substituting the NgoM IV-Cla I restriction fragment of the pACNR-17D-Rluc.2A-ic plasmid with the homologous fragment from pACNR-FLYF17Dx-ΔGDD, which contains a deletion in the cat-alytic GDD motif of 17D NS5 RNA dependent RNA polymerase domain.
-
pACNR-17D-Rluc.2A-ic, pACNR-17D-Rluc.2A-ic-ΔGDD and the corresponding SLA mutated plasmids were extracted by using PureLink HiPure Plasmid Midiprep Kit (Thermo Fisher Scientific) and linearized by Xho I diges-tion. The linearized DNA was used as templates for run-off in vitro transcription, which was performed as described previously (Liu et al. 2016) with minor modifications.
-
1.5 × 104 of BHK-21 cells were seeded into 48 well-plate the day before transfection. Until the cell confluency reached 60%–70%, 250 ng RNA per well was transfected into cells using Lipofectamine 2000 (Thermo Fisher Sci-entific) in triplicates following the manufacturer's instruc-tions. At the indicated time points, the supernatants of the transfected cells were collected for virus titer determination by plaque assay, and also served as the seed for recombi-nant virus amplification. The transfected cells were washed once with phosphate-buffered saline (PBS) and lysed with 1 × Renilla lysis buffer (Promega) and stored at - 20 ℃ for later Renilla luciferase assay.
-
For the RT-PCR identification of the recovered 17D-Rluc.2A, Viral RNA of 17D Rluc.2A (Passage 1, P1) and parental 17D was extracted using the E.Z.N.A. Total RNA Kit I (Omega). M-MLV reverse transcriptase (Takara) was used according to the manufacturer's instructions. The RT reactions were performed using YFV-R2 as the primer, and followed-up PCR amplifications were conducted with the primers YFV-F1 and YFV-R1. RT-PCR products were analyzed by electrophoresis on a 1% agarose/TAE gel.
To analyze the near full-length genome sequences of the recovered 17D-Rluc.2A virus and the corresponding SLA mutants, Total RNA was isolated from BHK-21 cells infected with the respective non-passaged viruses (P0). Then, reverse transcription was performed with primer YFV-R12 using Superscript III reverse transcriptase (Thermo Fisher Scientific). The cDNA products served as the templates for subsequent PCR reactions. Seven DNA fragments (S1 to S7) spanning the entire viral genome were amplified using the GoTaq Green Master Mix (Promega). DNA Sanger sequencing was performed by TSINGKE Biotechnology. The primers used for amplification and DNA sequencing were listed in Supplementary Table S1.
-
1 × 105 of BHK-21 cells were seeded into 48-well plates one day prior to infection. Cells were incubated with the corresponding virus at 0.1 multiplicity of infection (MOI) for 2 h followed by three times of washing with sterile PBS. Then the infected cells were cultured with fresh media containing 2% FBS. At the indicated time points, the supernatants were collected for titration by plaque assay.
-
The Renilla luciferase activity was measured by the Renilla luciferase assay system (Promega) using a Synergy H1 hybrid multi-mode microplate reader (BioTek, USA). In brief, 50-lL of 1 × Renilla luciferase substrate was auto-matically mixed with 20-lL of cell lysate in a F96 MicroWell black plate (Thermo Fisher Scientific), and signals were collected for ten seconds after a two-second incubation.
-
The P1 reporter virus was used as the start point for blind passage on BHK-21 cells. The supernatants were collected for further passage when a slight CPE appeared (typically 40–44 h after infection), and the cell lysates were collected for the reporter activity assay. In addition, viral RNA was extracted from the supernatant for RT-PCR and DNA sequencing to analyze the genetic stability of reporter virus as described before.
-
The templates for the in vitro transcription of the 5' -260 nt RNAs were prepared by high fidelity PCR using pACNR-17D-Rluc.2A-ic and the corresponding mutated plasmids as templates. The experimental procedures of in vitro transcription and SHAPE assay were described previously (Karabiber et al. 2013; Liu et al. 2016). The sequences for the extension primers were as follows: 5' -FAM-TGTTCTGGTCAGTTCTCTG-3' and 5' -HEX-TGTTCTGGTCAGTTCTCTG-3'. The fluorescent-labeled primer extension products were purified by ethanol/EDTA precipitation and resuspended with 10-lL of Hi-Di for-mamide (Thermo Fisher Scientific). Capillary elec-trophoresis analysis was performed by TSINGKE Biotechnology, and SHAPE data was analyzed by the Qushape software (Karabiber et al. 2013).
-
RNA structure prediction was performed by using the RNAstructure (Reuter and Mathews 2010) package version 5.8.1 and mfold (Zuker 2003). Secondary structures were drawn and annotated by using the VARNA software (Darty et al. 2009).
-
The compounds were twofold serially diluted in DMSO starting from 1 mmol/L for NITD008 (CAS number 1044589–82-3, Sigma) or 0.1 mmol/L for nanchangmycin (CAS number 35865–33-9, MCE). Experiments were per-formed in 48-well format. 1 × 105 of BHK-21 or A549 cells were infected with the indicated MOI of 17D-Rluc.2A under different compound concentrations. Cells incubated with medium containing the same concentration of DMSO (1%) served as the negative control. After culturing at 37 ℃ for the indicated hours, reporter activity in the infected cells were determined. Non-linear regression and the calculations of 50% effective concentration (EC50) were performed by using GraphPad Prism 7 (GraphPad Software). To evaluate whether the applied compound concentrations hindered cell viability, cells were seeded into 96-well plates and then incubated with different con-centrations of the compounds. At the indicated time points, 10-lL of CCK8 solution (Bimake) was added, followed by a one-hour incubation. Optical density (OD) values were measured using a Synergy H1 hybrid multi-mode microplate reader (BioTek, USA) at the 450 nm wave-length. To confirm the compounds did not affect the enzymatic activity of Renilla luciferase, various concen-trations of NITD008 or nanchangmycin were mixed into the cell lysates containing Renilla luciferase, and Renilla luciferase assay was performed.
-
The built-in statistical function of GraphPad Prism 7 was used for the correlation and linear/non-linear regression analysis. Two-way ANOVA and Bonferroni's multiple comparisons test were used to compare the titers of 17D-Rluc.2A and 17D. Two-way ANOVA and Tukey's multiple comparisons test were used to compare the expression levels of wild-type, ΔGDD and SLA mutant virus reporter genes. Other information about statistical analysis was listed in the corresponding figure legends.
Cell Culture and Virus
DNA Constructs
In vitro Transcription
Transfection of BHK-21 Cells with Reporter Virus RNA
RT-PCR and DNA Sequencing
Growth Curve Analysis
Renilla Luciferase Assay
Serial Passage and Genetic Stability Analysis
SHAPE Assay
RNA Structure Prediction and Demonstration
Antiviral Activity Assay
Statistical Analysis
-
To generate the 17D-Rluc.2A virus, the coding sequences of Rluc and FMDV 2A self-cleavage peptide were inserted after the 117th nucleotide of 17D C-coding region. This location was chosen to ensure the integrity of the identified and potential CREs in the first 1/3 of C-coding sequence (Liu et al. 2013; de Borba et al. 2015; Baker et al. 2020a). A partially re-coded full-length C gene was placed imme-diately after the 2A sequence (Fig. 1A, 1B). The aim of C gene-recoding is to abolish the redundant CREs involved in genome cyclization, such as the 5' cyclization sequence (5' -CS) (Bredenbeek et al. 2003), which may interfere with the correct mode of long-range RNA-RNA interactions. In vitro transcribed recombinant viral RNA was transfected into BHK-21 cells, and CPE was visible at 48 h post-infection (data not shown). Then the culture supernatant was collected and plaque assay was performed. Clear plaques formed after 4 days of incubation, suggesting that the 17D-Rluc.2A virus was successfully recovered. There was no apparent difference in plaque morphology between 17D-Rluc.2A and 17D, although the plaques formed by 17D-Rluc.2A were slightly smaller (Fig. 1C). To confirm the presence of the reporter gene in the recovered virus, viral RNA in the supernatants of 17D-Rluc.2A or 17D-infected BHK-21 cells were isolated and subjected to RT-PCR analysis. The PCR products were consistent with their expected length (Fig. 1D). DNA Sanger sequencing demonstrated that the inserted Rluc.2A sequence was intact. In addition, RT-PCR and DNA sequencing were performed to validate the genomic sequence of the recov-ered 17D-Rluc.2A and no unexpected mutations were found (data not shown). To further compare the biological property of 17D-Rluc.2A, a multi-step growth curve anal-ysis was performed in BHK-21 cells. It was shown that the 17D-Rluc.2A exhibited a statistically significant, but viro-logically moderate attenuation in propagation comparing with the parental 17D strain (Fig. 1E).
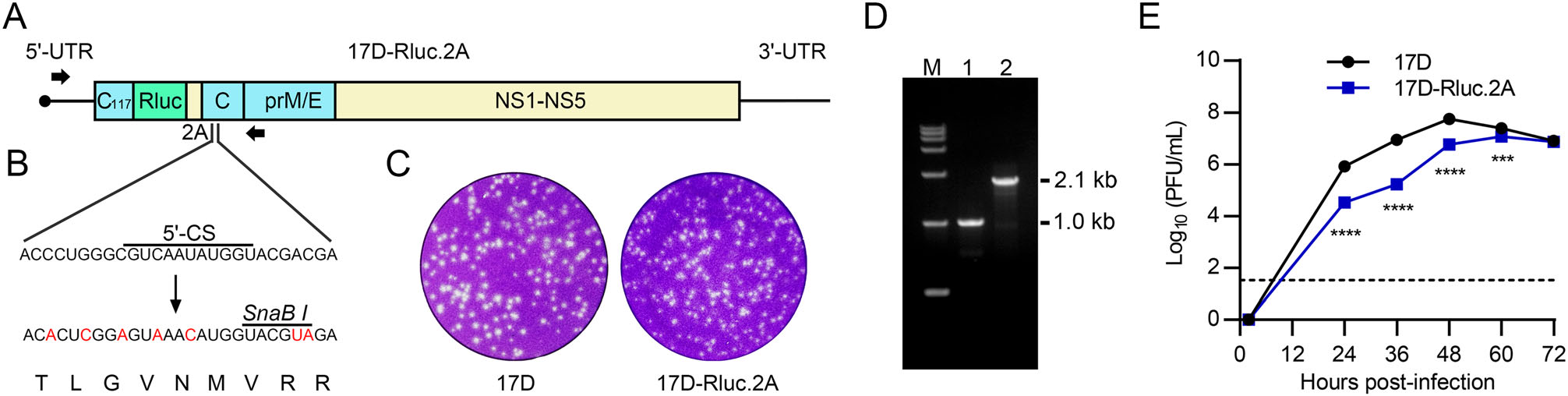
Figure 1. The construction and identification of the 17D-Rluc.2A reporter virus. A The genome organization of the 17D-Rluc.2A. The coding sequences of Rluc and 2A self-cleavage peptide were inserted after the 117th nucleotide in the C-coding region. B The full-length capsid gene downstream of 2A sequence was partially re-coded to abolish the redundant CREs, an artificial SnaB I restriction site was also introduced. C The plaque morphology of 17D-Rluc.2A and the parental 17D strain in BHK-21 cells. D RT-PCR characterization of the recovered virus. The primer binding region was shown in A as black arrows. The target sizes of amplification products were 1014 bp for 17D and 2136 bp for 17D-Rluc.2A. E Multi-step growth curves of 17D-Rluc.2A and 17D in BHK-21 cells. The detection limit is 33 PFU/mL. The experiment was performed in triplicates. Data was shown as means ± standard deviations in all figures. Two-way ANOVA and Bonferroni's multiple comparisons test were used to analyze statistical significance. ***P ≤ 0.001, ****P ≤ 0.0001.
-
The reporter gene expression in 17D-Rluc.2A-infected cells were then investigated. Firstly, BHK-21 cell lysates were collected at different time points after being trans-fected with 17D-Rluc.2A or the replication-lethal mutant 17D-Rluc.2A-ΔGDD RNA, and Renilla luciferase activity was determined. It was shown that the level of the reporter activity in 17D-Rluc.2A-transfected cells increased by more than 1, 000-folds from 6 to 48 h after transfection, whereas the reporter activity in the 17D-Rluc.2A-ΔGDD group gradually reduced, suggesting that the reporter expression depended on viral replication (Fig. 2A). Effi-cient reporter expression was also observed in infected C6/36 cells (Fig. 2B). Secondly, the supernatants and cell lysates of transfected BHK-21 cells were collected at 24, 36 and 48 h post-infection, and subjected to plaque assay and luciferase assay respectively. By comparison of the intracellular reporter expression and extracellular progeny viral load, we found a good linear correlation between the reporter expression level and progeny virus production in the logarithmic form (Fig. 2C), suggesting that the reporter expression level of 17D-Rluc.2A is a faithful indicator of viral replication. At last, the stability of the reporter expression after serial passage of 17D-Rluc.2A was investigated. We found that the reporter expression level only moderately reduced during passage 2 to passage 3, whereas an apparent decrease of reporter level occurred at passage 4 (Fig. 2D). RT-PCR and DNA sequencing anal-ysis of the passaged viral RNA confirmed that the inserted fragment was gradually lost, suggesting that 17D-Rluc.2A is genetically unstable after extended passages. For this reason, only 17D-Rluc.2A stocks within two passages were used in the subsequent assays.
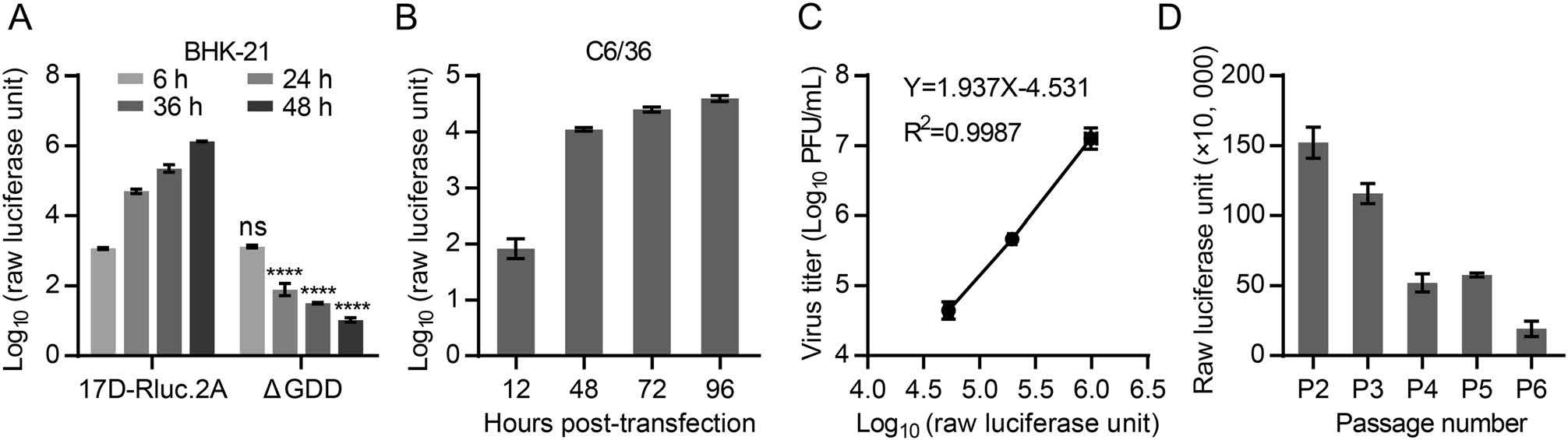
Figure 2. 17D-Rluc.2A infection leads to robust reporter gene expression in both mammalian and mosquito cells. A The expression of the Renilla luciferase in the 17D-Rluc.2A-transfected BHK-21 cells. The 17D-Rluc.2A-ΔGDD RNA, which containing a deletion of the catalytic GDD motif in NS5 RNA-dependent RNA polymerase, was transfected in parallel as a negative control. The reporter levels between 17D-Rluc.2A-ΔGDD and 17D-Rluc.2A at the same time points were compared. Two-way ANOVA and Tukey's multiple comparisons test were used to analyze statistical significance. ns: P > 0.05, ****P ≤ 0.0001. B The expression of Renilla luciferase in 17D-Rluc.2A-infected C6/36 cells (MOI = 0.1). C The correlation between intracellular reporter activity and extracellular virus titer in 17D-Rluc.2A-transfected BHK-21 cells. A regression equation was listed. Y = Virus titer (Log10 PFU/mL), whereas X = Log10 (Raw luciferase unit). D Monitoring of reporter luciferase activity during the serial passage of 17D-Rluc.2A. The experiments were performed in triplicates.
-
To demonstrate that 17D-Rluc.2A can be utilized to investigate the mechanism of YFV replication, we introduced mutations into a highly-conserved flavivirus CRE, the 5' -SLA (Fig. 3A), which is crucial for the recruitment of NS5 to viral RNA in dengue virus (Filomatori et al. 2006). The SLA-M1 mutation was aimed to disrupt the base-pairing of the stem 2 motif in 5' -SLA, whereas the SLA-M2 mutation restored the corresponding base-pairing. The RNA transcripts of wild type (WT), ΔGDD or SLA-mutated 17D-Rluc.2A were transfected into BHK-21 cells. At 6 h after transfection, the reporter activities in the transfected cells were similar (data not shown), suggesting that the SLA mutations do not interfere with viral translation apparently. As expected, the lucifer-ase activity of the WT group (showed as relative luciferase unit) increased rapidly at 6 h to 48 h post-transfection and those values of the ΔGDD group decreased over time (Fig. 3B). The relative luciferase unit of the SLA-M1 group gradually dropped after transfection and the overall reporter expression tendency was similar between the ΔGDD and SLA-M1, suggesting that the disruption of SLA stem 2 was lethal to YFV. In comparison, the replication profile of the SLA-M2 mutant was apparently restored (Fig. 3B). To better analyze the effect of SLA mutation on viral replication using the reporter virus, BHK-21 mono-layers were incubated with culture supernatants of the WT, SLA-M1 or SLA-M2-transfected cells, and the reporter expression levels were monitored (Supplementary Fig. S1). It was shown that the differences in relative luciferase unit were similar with the results in Fig. 3B, and the extremely low luciferase signal in SLA-M1 blind-infected cells con-firmed that the SLA-M1 mutant is lethal. Moreover, sequencing analysis confirmed that the designed mutations were present in the recovered SLA-M2 virus, with no additional mutations found (data not shown). Interestingly, the SLA-M2 consistently replicated less efficiently than the WT during the early stage of viral replication, agreeing with our previous results in Zika virus (Liu et al. 2017). Next, to confirm the effect of SLA mutation on the sec-ondary structure of 5' -SLA, SHAPE assay was performed for either WT, SLA-M1 or SLA-M2 mutated YFV 5' -260 nt RNA (Fig. 3C). The SHAPE reactivity pattern of the SLA-M1 was apparently altered comparing with the WT, whereas the SLA-M2 had a SHAPE reactivity pattern resembling that of the WT. Then RNA secondary structure prediction was performed by using SHAPE reactivity as constraints (Fig. 3D). We found that the SLA-M1 mutation caused a refolding of the entire 5' -SLA, which may con-tribute to its lethal phenotype. The results of RNA structure prediction also confirmed the restoration of 5' -SLA struc-ture by the complementary SLA-M2 mutation. Taken together, these results demonstrated that the replicational phenotype of 17D-Rluc.2A 5' -SLA mutants agreed well with the proposed function of this CRE and in vitro bio-chemical results, suggesting that 17D-Rluc.2A can be used for the study of YFV replication.
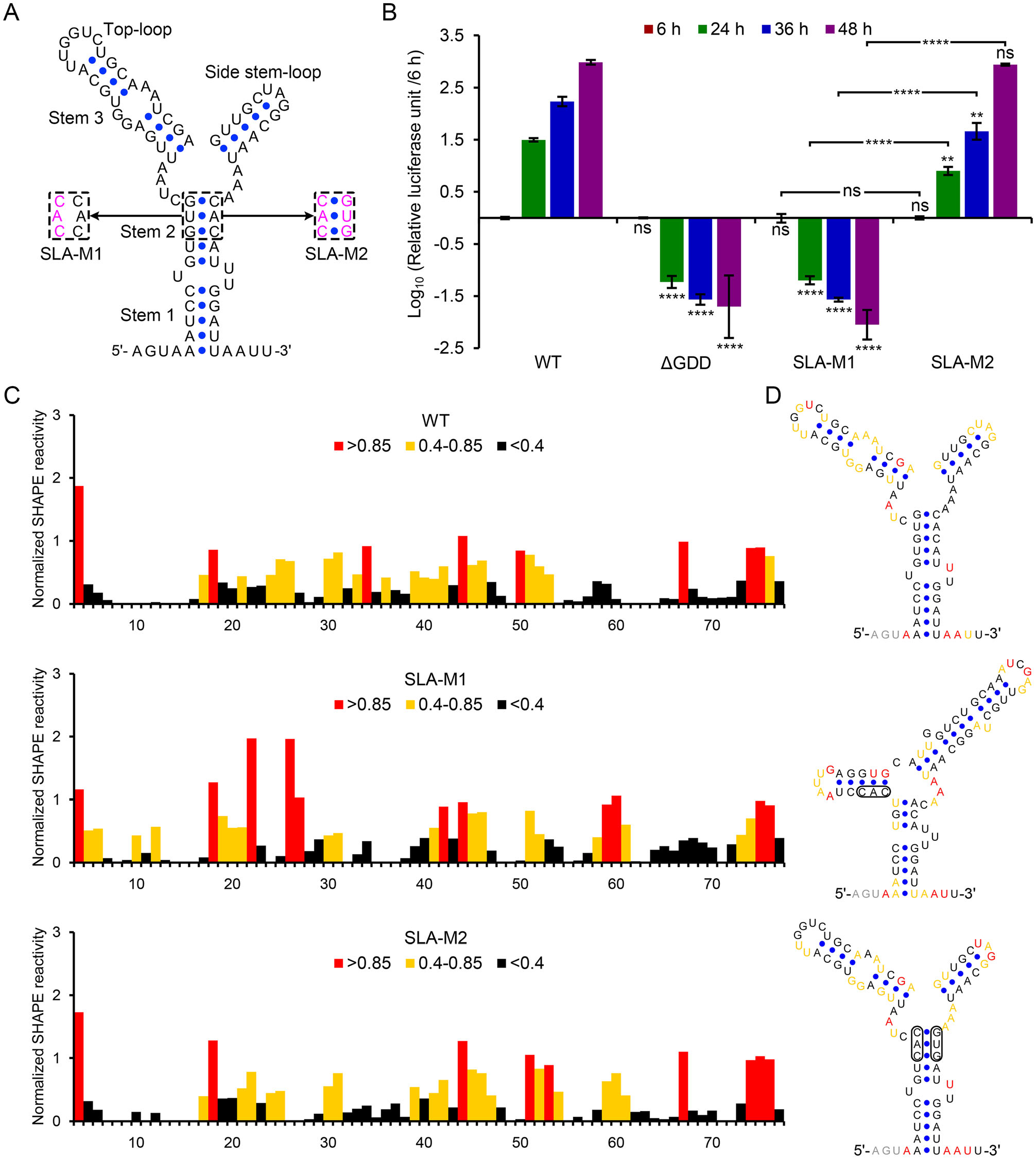
Figure 3. The function of 5' -SLA in yellow fever virus replication analyzed by using 17D-Rluc.2A. A The secondary structure of YFV 5' -SLA and mutation design. B the reporter activities in WT, ΔGDD or 5' -SLA mutant RNA-transfected BHK-21 cells were measured at different hours post-transfection. Relative luciferase unit (the ratio of the reporter activity measured at later time points to the value measured at 6 h post-transfection) was shown in logarithmic form. The experiment was performed in triplicates. Two-way ANOVA and Tukey's multiple comparisons test were used to analyze statistical significance. Unless indicated by connecting lines, P values referred to the comparisons of the logarithmic relative luciferase units between the ΔGDD, SLA-M1 or SLA-M2 group and the WT group at the same hours post-transfection. ns: P > 0.05, **P ≤ 0.01, ****P ≤ 0.0001. C SHAPE analysis of the 5' -SLA region of the WT and mutants. D SHAPE reactivity-guided structure prediction of WT and mutated 5' -SLA sequences.
-
To assess the potential application of 17D-Rluc.2A in the evaluation and screening of anti-viral medicines, NITD008 (Yin et al. 2009), an inhibitor of flavivirus RdRp was firstly utilized. To this aim, we infected BHK-21 or A549 cells with 0.1 MOI of 17D-Rluc.2A in the presence of two-fold diluted NITD008 or solvent DMSO only, and the reporter activity of infected cells was measured at 48 h post-infection (Fig. 4A), when viral replication almost reached the peak. As expected, NITD008 inhibited reporter activity in 17D-Rluc.2A-infected cells in a dose-dependent manner, and the calculated EC50 was 1.06 lmol/L in BHK-21 cells and 0.26 lmol/L in A549 cells (Fig. 4B, 4C). To exclude the possibility that NITD008 interfered with cell physiology, CCK8 assay was performed. No detectable cytotoxicity caused by NITD008 at the corresponding EC50 concentra-tions was observed. Additionally, direct addition of NITD008 into control luciferase activity assays indicated that it did not interfere with the reporter assay per se (data not shown). Thus, the anti-viral activity assay based on the reporter virus is specific and reliable. We then investigated whether a recently reported inhibitor of Zika virus, nan-changmycin (Rausch et al. 2017; Hackett and Cherry 2018) also inhibits YFV infection. Since nanchangmycin was reported to inhibit the internalization of several flaviviruses, the anti-viral activity assay was modified accordingly. BHK-21 or A549 cells were infected with 0.01 MOI of 17D-Rluc.2A, and the reporter activity was measured at 24 h post-infection (Fig. 4D). These modifications were aimed to slow down the dissemination of virus between cells so that the entry process is still occurring at the time point of detection. A robust inhibitory effect of nanchangmycin on the reporter expression in 17D-Rluc.2A-infected BHK-21 (EC50 = 0.069 lmol/L) or A549 cells (EC50 = 0.055 lmol/L) were detected (Fig. 4E, 4F), and this effect was not caused by cytotoxicity or inhibition of luciferase activity (data not shown). The calculated EC50 values of NITD008 and nan-changmycin herein were similar to those in previous reports for other flaviviruses (Yin et al. 2009; Deng et al. 2016; Lo et al. 2016; Rausch et al. 2017). These results demonstrated that the 17D-Rluc.2A is a potent tool to evaluate the therapeutic potentials of future anti-YFV medicine.

Figure 4. The evaluation of antiviral efficacy of NITD008 and nanchangmycin based on 17D-Rluc.2A. A Experimental protocol to test the anti-YFV activity of NITD008. B–C The inhibition of 17D-Rluc.2A propagation by NITD008 was monitored by the reporter activity in BHK-21 (B) or A549 cells (C), cells incubated with medium containing 1% DMSO served as the negative control. MOI in (B) and (C) was set to 0.1. D Experimental protocol to test the antiviral activity of nanchangmycin. E–F. The inhibition of 17D-Rluc.2A infection by nanchangmycin as monitored by the reporter activity in BHK-21 (E) or A549 cells (F), cells incubated with medium containing 1% DMSO served as the negative control. MOI in (E) and (F) was set to 0.01. The experiments were performed in triplicates. R-square values of the regression and EC50 values (defined as 50% reduction of the raw luciferase unit) were presented.
Generation of the 17D-Rluc.2A reporter virus
Characterization of the Reporter Gene Expression of 17D-Rluc.2A
Characterization of the 5' -SLA Element Using 17D-Rluc.2A
Identification of Anti-YFV Compounds Using 17D-Rluc.2A
-
The development of reporter flaviviruses has greatly advanced in recent years (Zhang et al. 2016, 2019; Kassar et al. 2017; Baker et al. 2020a, 2020b; Baker and Shi 2020). In this work, we utilized the well-established 2A self-cleavage peptide strategy to generate the 17D-Rluc.2A reporter virus. The in vitro phenotypes of 17D-Rluc.2A shared similarity to the parental 17D strain, and the reporter expression in the infected cells correlated well with progeny virus production, indicating the successful generation of a reporter virus.
We then investigated the function of the conserved 5' - SLA element in YFV replication based on 17D-Rluc.2A. The results confirmed the expected crucial role of 5' -SLA in YFV life-cycle, indicating that the 17D-Rluc.2A is suitable for the investigations of YFV replication mecha-nisms. In combination with SHAPE technology, we found that there were still regions with differential SHAPE reactivities between SLA-M2 and WT (Fig. 3C, 3D). Notably, the SHAPE reactivities of the SLA-M2 top-loop were quite different from those of the WT, suggesting that possible tertiary interactions exist within the 5' -SLA. Since the top-loop is an important functional motif in 5' -SLA (Lodeiro et al. 2009; Liu et al. 2017), these differences in SHAPE reactivity may reflect some alternation in its 3-D conformation induced by changes in the base-pair com-position of the SLA-M2 mutant, which probably accounts for the moderately attenuation of the SLA-M2.
Several reporter YFV have been generated (Kassar et al. 2017; Baker et al. 2020a; Baker and Shi 2020; Sanchez-Velazquez et al. 2020; Syzdykova et al. 2021) for different research interests. Based on their design, these reporter viruses can be classified into two groups. In yellow fever reporter viruses like YFV-GLuc (Kassar et al. 2017), YFVhb (Sanchez-Velazquez et al. 2020) and pCMV-YFV/GFP-NS1 (Syzdykova et al. 2021), the reporter genes were either inserted between the E and NS1 coding region, or fused with the NS1 gene. In a recently reported yellow fever reporter viruses expressing the NanoLuc luciferase (Baker et al. 2020a, 2020b) and the reporter virus described herein, the reporter genes were inserted near N-terminal of the viral polyprotein. Comparing with the YFV-GLuc, YFVhb and pCMV-YFV/GFP-NS1, 17D-Rluc.2A is more suitable for the research of CREs, especially those in the C-coding region, since these CREs were successfully uncoupled from C-coding sequence in 17D-Rluc.2A. On the other hand, compared with the NanoLuc-based reporter YFV, our reporter virus is cost-effective, which is an important factor of consideration for high-throughput screening. Thus, the reporter virus reported herein will be of great value for both fundamental studies and drug development of YFV.
-
The authors thank for Professor Charles M. Rice from Rockefeller University for the kindly sharing of the infectious clone of 17D vaccine strain. We are grateful for the helpful discus-sions with Professor Pei Xu of School of Medicine, Sun Yat-sen University, and Professor Guang-Chuan Wang of the Center for Excellence in Molecular Cell Science, CAS. We thank for Professor Hua Zhang, School of Medicine, Sun Yat-sen University for the providing of the A549 cell line. This work is supported by the National Natural Science Foundation of China (81871632 and 32070183), the Natural Science Foundation of Guangdong Province (2020A1515010656), and the Creative Research Group Foster Project of the Sun Yat-sen University. Professor Zhong-Yu Liu is supported by the One-Hundred People Project of the Sun Yat-sen University.
-
ZYL proposed the project. ZYL and DL designed the experiments. DL and JLY performed the experiments. ZYL and DL analyzed and interpreted the data. DL and ZYL wrote the manuscript. All authors read and confirmed the submission of the paper.
-
The authors declare that they have no conflict of interest.
-
This article does not contain any studies with human or animal subjects performed by any of the authors.
















 DownLoad:
DownLoad: