HTML
-
Baculoviruses are large double-stranded DNA viruses of invertebrates. The type species Autographa californica nucleopolyhedrovirus (AcMNPV) encodes 154 proteins, including much of its own DNA replication and transcription machinery. Baculoviruses use the host RNA polymerase for transcription of the early genes, but encode their own RNA polymerase for the synthesis of late and very late genes. While viral RNA polymerases are common in virology, baculoviruses provide the only known example of a eukaryotic virus that replicates in the nucleus and yet duplicates the host transcription machinery. One consequence of using a viral RNA polymerase is the need to also provide unique solutions for RNA processing. This review summarizes what is known regarding the mechanisms of RNA processing in baculovirus-infected cells. This knowledge primarily stems from research on AcMNPV, but should be applicable to all members of the family.
Vlak et al. (31) were the first to demonstrate that late mRNAs were polyadenylated at their 3' ends. They used oligo (dT) to purify adenylated RNAs from infected cells that were harvested at late times of infection. In vitro translation of polyA RNA produced viral structural proteins including capsid and polyhedrin. One year later, Qin and Weaver (25) examined the 5' ends of viral mRNAs. They also purified RNA from the late phase of infection and showed that the bulk of the polyadenylated RNAs had cap1 structures (m7GpppAm), consisting of a methyl group on the N7 of the guanine cap and a methylated 2'-O on the ribose of the first transcribed base.
At the time of these discoveries, the mechanisms by which RNA processing enzymes recognize their substrates was unknown. Most of the baculovirus community assumed that processing of viral RNAs was mediated by the same enzymes that catalyzed these modifications of the host mRNAs. At least for polyadenylation, this assumption was supported by the presence of appropriately spaced cleavage/polyadenylation signals (AATAAA) in the 3' untranslated region of most viral late genes. A study showing that a synthetic globin cleavage/polyadenylation cassette could substitute for the native 3' non-coding region of the polyhedrin gene further cemented the idea that host enzymes were involved in 3' processing (32). As a result, many baculovirus expression vectors were developed with the SV40 cleavage/polyadenylation signal sequences in place of polyhedrin downstream sequences (17).
In the late 1990s, research on cellular mRNA processing enzymes revealed the mechanism by which these enzymes recognize their substrates (3, 21, 22). The major determinant for processing is a not feature that is intrinsic to the RNA, but is merely a consequence of their transcription by cellular RNA polymerase Ⅱ. Even the cleavage/polyadenylation signal is not sufficient for 3' end processing. It is important in positioning the complex for cleavage, but is only recognized in the context of RNA polymerase Ⅱ. The polymerase feature that is recognized by mRNA processing enzymes is the C-terminal domain (CTD). This structure is not found on the other cellular polymerases and is thus unique to RNA polymerase Ⅱ. The capping and cleavage/polyadenylation enzymes interact with the CTD and process all RNAs that are transcribed by that enzyme.
Identification of the baculovirus RNA polymerase in 1998 revealed that it was a complex of 4 viral subunits (8). LEF-8 and LEF-9 likely comprise the catalytic domain of the viral RNA polymerase. LEF-8 is a 102-kDa protein with a conserved 13-amino acid sequence motif that is found in the β' family of DNA-directed RNA polymerases. This corresponds to conserved block H in E.coli RNA polymerase, and structural studies indicate that this region forms part of the catalytic site (24, 27). LEF-9 contains an aspartic triad (NADFDGD), which is common to the β family of RNA polymerase subunits. As discussed below, the LEF-4 subunit is an mRNA capping enzyme (5, 6, 10). The specific role of P47 in late transcription is unknown. Only LEF-4 can be expressed as a single subunit, thus limiting the biochemical characterization of subunit functions.
Identification of the RNA polymerase subunits invited a fresh examination of RNA processing in baculovirus infected cells. Significantly, none of the RNA polymerase subunits have a sequence resembling the CTD that could potentially interact with the mRNA processing enzymes. This suggested that baculoviruses would probably encode their own enzymes for capping and polyadenylation.
-
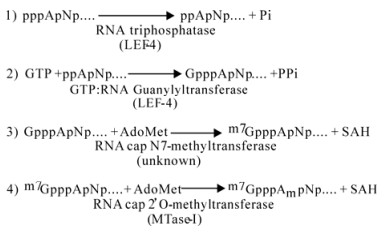
Formation of a cap1 structure requires four different enzymatic activities (Fig. 1). The first is RNA 5'-triphosphatase. This enzyme removes the gamma phosphate from triphosphate-terminated nascent strands to produce a disphosphate-terminated RNA. The second enzyme is GTP: RNA guanylyltransferase. This transfers GMP from GTP to the diphosphate ends, forming the inverted GpppN cap. The guanylation reaction is freely reversible until the cap is stabilized by methylation of guanine at N7 to form the cap0 structurem7 GpppA. This methylation reaction is mediated by the enzyme RNA cap N7-methyltransferase. Methylation of the 2'-OH on the ribose of the first transcribed base produces the cap1 structure. This second methylation is catalyzed by RNA cap 2'O-methyltransferase.
The enzyme that catalyzed the first two reactions was identified by two groups using different approaches. We reasoned that the capping enzymes should associate with the baculovirus RNA polymerase (6). Therefore, we monitored guanylyltransferase activity throughout the purification of baculovirus RNA polymerase. This is a two-step reaction involving the formation of a covalent enzyme-guanylate intermediate in which GMP is linked to a lysine residue in the enzyme (29). The intermediate can be conveniently assayed by 32P label transfer from [α-32P] GTP to the enzyme. Subsequent analysis of enzyme-label (E*pG) complex reveals the size of the guanylyltransferase, making its identification relatively simple. We found that guanylyltransferase activity tightly copurified with RNA polymerase. Furthermore, even the foursubunit complex had activity, and radiolabel was associated with the LEF-4 subunit of viral RNA polymerase (Fig. 2). Expression of the single subunit in bacteria confirmed that it had guanylyltransferase activity, while a mutant version with a substitution in the essential KxDG motif was not able to bind GMP.
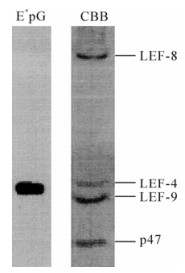
Figure 2. Identification of LEF4 as a capping enzyme. Purified RNA polymerase was incubated with [α-32P] GTP and separated on SDS-polyacrylamide gels. The left lane shows the migration of protein radiolabeled with 32P (E*pG) and the right lane shows the Coomassie brilliant blue (CBB) stained polymerase complex. The identity of the four subunits is indicated on the right.
Gross and Shuman looked specifically for guanylyltransferase motifs in the RNA polymerase subunits (5). Although the extent of sequence conservation among these proteins is not extensive enough for computer alignment programs, sequence gazing revealed motifs that were common among guanylyltransferases of poxviruses and yeast and baculovirus LEF-4 proteins.
After mapping guanylyltransferase activity to the C-terminal half of LEF-4, it was an obvious guess that RNA triphosphatase activity would also map to N-terminal portion of LEF-4 (5, 10). This supposition was based upon a similar arrangement in the vaccinia virus capping enzyme, and the conservation of two glutamic acid dyads that are essential for function in vaccinia capping enzyme (34). Both the LEF-4 single subunit and the 4-subunit RNA polymerase cleaved gamma phosphates on triphosphate-terminated RNA. In addition, both enzymes released gamma phosphates from free NTPs. The RNA triphosphatase activity required a metal cofactor, either manganese or magnesium. NTP hydrolysis, however, was activated only by manganese.
Mutagenesis studies showed that the RNA triphosphatase and guanylyltransferase domains of LEF-4 functioned independently (10), and that the N-terminal 236 amino acids could function as an autonomous triphosphatase domain (18).
The lef-4 gene was first identified as a late expression factor that was required for transient expression of a reporter construct under the control of a baculovirus late promoter (23). A total of 19 factors are required for late expression by this assay, including the other three subunits of RNA polymerase (26). The transient expression data and the identification of LEF-4 as an RNA polymerase subunit were consistent with ongoing studies of a lef-4 temperature-sensitive (ts) mutant (1). The phenotype of this mutant indicated a deficiency in late gene expression at the nonpermissive temperature. The ts lesion was mapped to a single nucleotide transversion that resulted in the substitution of phenylalanine for leucine at amino acid residue 105. Although this residue is within the RNA triphosphatase domain, protein activity and stability studies showed that the mutant protein was not deficient in RNA triphosphatase activity (10). This raised the possibility that LEF-4 might have yet another function in late transcription, most likely as an essential structural component of the viral RNA polymerase.
To further analyze the essential function of LEF-4, we performed RNA silencing experiments and gene knock-out experiments using bacmid technology (13). RNA silencing confirmed that the absence of LEF-4 specifically repressed the expression of late genes, without affecting the synthesis of early genes, thus confirming that essential functions of LEF-4 were limited to late gene expression. Construction of a bacmid virus containing an insertion in the lef-4 gene confirmed the lack of replication due to the absence of this essential gene. Furthermore, the availability of a knockout virus provided a method to analyze the ability of specific mutant versions of lef-4 to rescue the phenotype. Co-transfection with a plasmid encoding wildtype LEF-4 rescued the impaired virus, but transfection with a guanylyltransferase mutant did not. This confirmed that capping is an essential function of LEF-4, and further suggests that the ts virus lesion may be due to loss of integrity in the polymerase complex. Studies concerning whether the RNA triphosphatase function of LEF-4 is essential for viral infection are ongoing in my laboratory (Li and Guarino, unpublished).
-
Baculoviruses also encode another protein with RNA triphosphatase activity. This protein was originally named PTP because of its homology with members of the protein tyrosine phosphatases, as well as its ability to dephosphorylate serines, threonines and tyrosines on protein substrates (28). It was subsequently recognized that proteins in this subfamily, containing a signature HCTHGxNRT motif function, as RNA 5'-triphosphatases in RNA capping (4, 30). This family of capping enzymes is found primarily in metazoans, while DNA viruses, protozoa and yeast, use the LEF-4 type enzyme. The two types of RNA triphosphatases catalyze the same reaction, but are structurally and mechanistically distinct (2, 19). The viral enzymes use metal cofactor that is coordinated by essential glutamates, while the metazoan enzymes are metal-independent and act via a phosphocysteine intermediate.
While there is no evidence that BVP is involved in mRNA capping, studies with the closely related Bombyx mori nucleopolyhedrovirus (BmNPV) revealed that viruses with knockout mutations in the gene have a very interesting phenotype (12). They are deficient in wandering, which is a behavioral response to baculovirus infection. Insects climb to the top of plants or trees on which they are feeding shortly before death. This is advantageous to virus dissemination because virions are released on the young tender leaves where insects are most likely to feed. The role of BVP in this complex behavioral pathway is not understood, but the involvement of an RNA processing enzymes suggest that regulatory RNA is involved at some step of the process.
Baculovirus BVP differs from cellular RNA triphosphatases in that it also removes β phosphates from RNAs and free NTPs (4). This activity would seem to suggest that BVP would not make a good RNA capping enzyme because β phosphate hydrolysis would preclude the second step in capping. Experiments by Martins et al., (20), however, demonstrated that bvp could complement a lethal deletion of the yeast RNA triphosphatase. Presumably it could function in this capacity because enzyme action is distributive, allowing the diphosphate to be capped before it was hydrolyzed. This result indicates that BVP may be involved RNA capping in baculovirusinfected cells, although this activity would be redundant to LEF-4. Furthermore, the fact that lef-4 is an essential gene, while bvp is dispensable (15) suggests that it would be an ancillary role in capping, if any.
-
The enzyme that catalyzes the third step in cap formation (guanine-N7 methylation) has not yet been identified. The AcMNPV genome encodes only one protein with predicted methyltransferase activity. This protein, named MTase1 (ORF69), belongs to the RNA cap 2'O-methyltransferase family, based on conserveation of certain residues that interact with RNA substrates. Furthermore, biochemical studies demonstrated that it bound the cofactor S-adenosyl-methionine and methylated capped RNAs (33). Unlike other members of RNA cap 2'O-methyltransferases, methylation of the ribose was not dependent upon prior methylation at N7. This indicates that methylation of the ribose could occur either before or after the guanine cap methylation.
MTase1 is not highly conserved among baculoviruses. It has been found in most, but not all of the Group 1 NPVs and approximately half of the Group 2 NPVs. Thus far, there have been no reports of MTase1 orthologs in the GVs or in the dipteran or hymenopteran NPVs. This lack of absolute conservation is consistent with data showing that mtase 1 is not an essential gene in AcMNPV, although deletion of it decreased the yield of progeny virus production (33). Transient assay data, however, showed that mtase 1 increased expression of reporter genes under the control of a late viral promoter (14). Therefore, mtase 1 could also be classified as an accessory LEF. This would be consistent with its role in cap formation, as this is known to stabilize RNAs and should contribute to their translation.
The finding that mtase 1 stimulated transient late gene expression, but was dispensable for viral replication may seem contradictory. Similar results have been noted for at least two other lef genes, namely lef-12 and lef-6 (7, 16). This suggests that other viral proteins may compensate for their absence.
Given the presence of LEF-4-encoded RNA triphosphatase and guanylyltransferase in all baculoviruses and 2'O-methyltransferase in most of the lepidopteran NPVs, it is somewhat surprising that the virus does not encode a good candidate for RNA cap N7-methyltransferase. It is possible that baculoviruses contain another methyltransferase that has low sequence conservation and has not been identified. Alternatively, host enzymes could be recruited for guanine N7 methylation. Preliminary results from our lab (Wu and Guarino, unpublished) indicate guanine N-7 methyltransferase activity increases after infection, consistent with the idea that the virus encodes this activity or at least stimulates the host enzyme. Given that this modification is essential for cap stability, it is not surprising that the virus regulates enzymatic activity in some manner.
LEF-4
PTP/BVP
MTase-1
-
To determine whether a cellular cleavage/polyadenylation signal could direct baculovirus 3'end formation, Westwood et. al. inserted a synthetic globin sequence into the polyhedrin locus (32). This sequence contained the two major determinants of a cleavage/polyadenylation signal: a hexanucleotide AAUAAA and a downstream GU-rich sequence. Experimental data revealed that approximately half of the transcripts initiating at the polyhedrin promoter had 3' ends consistent with termination and polyadenylation within the globin sequence. The other transcripts read through this sequence and their 3' ends were processed at the native polyhedrin locus. The relatively high efficiency of the globin sequence suggested to many researchers that baculovirus 3' ends were formed by the same mechanism as host RNAs, and was most likely mediated by host enzymes.
Subsequent research on the mechanism of cellular mRNA polyadenylation revealed that the cleavage/ polyadenylation machinery identified their substrates through interactions with the CTD of RNA polymerase Ⅱ. Furthermore, RNA polymerase Ⅱ was found to be an essential factor in the polyadenylation step (9). After the identification of a baculovirus RNA polymerase lacking a CTD, we decided to revisit the question of whether host enzymes were responsible for mRNA processing at the 3' end (11). We noted that the globin signal sequence used by Westwood et al. contained a U-rich sequence, and predicted that it would serve as a termination signal. Baculovirus 3' non-coding regions are very A+T-rich and contain multiple iterations of U-rich sequences as well as the canonical AATAAA cleavage/polyadenylation sequence.
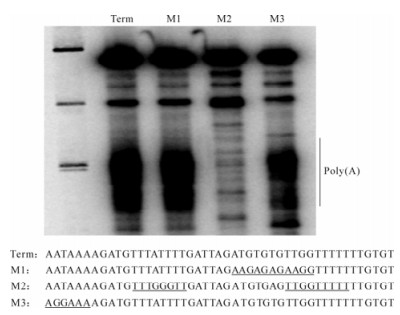
To test whether the globin cleavage/polyadenylation signal was recognized by baculovirus RNA polymerase as a termination signal, it was cloned into the transcription template Polh/CFS, which directs synthesis of RNA under the control of the polyhedrin promoter. We predicted that transcripts from Polh/ CFS-Term would be 47 nt longer than those obtained with the parental construct if the globin cassette was not recognized (Fig. 3C). Alternatively, they would be approximately 69 nt transcription shorter if they were terminated or cleaved within the globin sequence. In vitro assays revealed the presence of both longer and shorter transcripts (Fig. 3A). This result agreed with those of Westwood et al., indicating that the efficiency of the globin sequence was approximately 50%. The mechanism of 3' end formation, however, was shown to be termination followed by polyadenylation and not cleavage/polyadenylation. Two lines of evidence supported this. First, a time course showed that the fulllength product was stable, suggesting that it was not subject to cleavage (Fig. 3B). During this time, the heterogeneity of the shorter transcript increased, consistent with polyadenylation of released terminated transcripts. 3'RACE analysis of the transcripts confirmed that they were polyadenylated after transcription of oligoU. Secondly, mutational analyses showed that neither the AAUAAA nor the GU-rich sequence were required for termination or polyadenylation by viral RNA polymerase (Fig. 4). The U-rich sequence, however, was essential. The fact that the four-subunit RNA polymerase was capable of polyadenylating the terminated transcripts suggests that a non-templated nucleotide synthesis activity is intrinsic to the enzyme, although this has not been experimentally determined. The construction of mutant versions of polymerase that are unable to synthesize RNA to polyadenylated would help to determine whether the same active site is used for both activities.
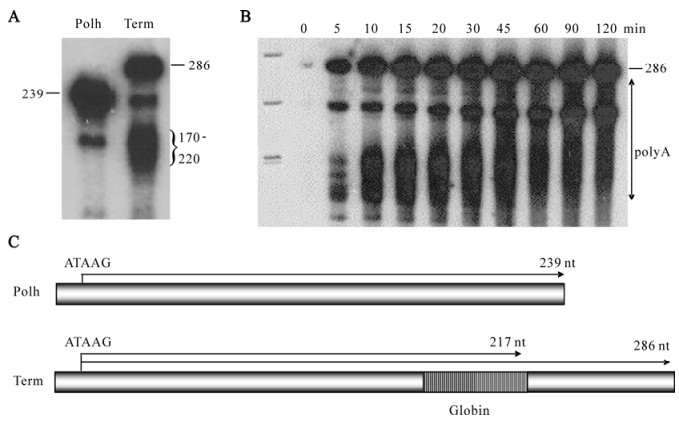
Figure 3. Intrinsic termination and polyadenylation activities of baculovirus RNA polymerase. A: Transcripts produced by RNA polymerase on the standard template containing the polyhedrin promoter (Polh) or the standard template with the globin cleavage/polyadenylation cassette (Term). The sizes of the products are labeled on the left or right, respectively. B: Time course of synthesis. The incubation times are indicated on the top and the sizes of the full-length product and the range of polyadenylated products is indicated on the right. C: A schematic of the templates.
-
Unlike all other DNA viruses that replicate in the nucleus of their host cells, baculoviruses encode their own DNA-directed RNA polymerase to specifically transcribe and process late and very late messages.
This polymerase is a remarkable enzyme that has the ability to recognize viral promoters, transcribe linked genes, cap the 5' ends of the RNA, terminate synthesis at oligoU-rich regions, polyadenylated the released transcripts. There is minimal sequence similarity between the baculovirus RNA polymerase and other viral and cellular polymerases and processing enzymes. Further study of the baculovirus RNA polymerase and accessory factors will surely reveal novel mechanisms for the regulation of viral gene expression.













 DownLoad:
DownLoad: