-
Severe acute respiratory syndrome (SARS) is caused by a novel coronavirus called SARS-CoV (7, 8, 10). The virus causes atypical pneumonia with diffuse alveolar damage with an overall mortality of 10% that ranges from 0% in children and 50% in persons over 65,seriously threatening public health worldwide.
The SARS-CoV is an enveloped virus of a positive single stranded RNA genome~29.7 kb in length and has 11 open reading frames. The genome organization is similar to that of other coronavirus,but phylogenic analyses and sequence comparisons showed that SARS-CoV is not closely related to any of the previously characterized coronavirus (7, 10). The structural protein genes of SARS-CoV are contained in four major open reading frames,and include the surface spike(S) glycoprotein,the small membrane protein(M),the envelope(E) glycoprotein,the nucleocapsid(N) protein,and a set of accessory proteins whose number and sequence vary among different coro-naviruses (11).
The S protein of SARS-CoV is a large type Ⅰ membrane glycoprotein projection from the viral envelope which is responsible for both binding to receptors on host cells and for membrane fusion. Angiotensin converting enzyme 2(ACE2) was found to be an efficient receptor for the S glycoprotein of SARS-CoV (9, 4, 12). The S protein also contains important virus neutralizing epitopes that elicit neutralizing antibody in the host species (3, 2). To determine the critical sequence of the S protein that interacts with the ACE2 receptor,the interactions between ACE2 and different truncated S proteins were investigated. Our study sheds some light on the interaction mechanism and provides useful insight into the development of a protein vaccine for SARS-CoV.
HTML
-
Plasmid pGEM-S encoding SARS-CoV S protein was provided by Prof Zhu Ying in the State Laboratory of Virology,College of Life Science in Wuhan University. Prokaryotic expressing vectors pET-His were purchased from Gene Power Lab Ltd.,Shenzhen office. E.coli BL21(DE3) and DH5 were from our laboratory. T4 DNA Ligase was from TaKaRa,DNA restrictive endoenzymes were from Jingmei Biotech.Plasmid pProEXHTb vero ACE2 and the rabbit anti sera against ACE2: Anti veroACE2 was provided by Dr. Hu Zhi Hong of Wuhan Institute of Virology,Chinese Academy of Sciences.
-
Plasmids S1-1 encoding SARS-Cov S protein residues 262~606,S1-2 encoding 314~606,S1-3 encoding 388~606,S1-4 encoding 314~387,S1-5 encoding 262~496,S1-6 encoding 262~387,S1-7 encoding 388~496 (Fig. 1.) were constructed. The sequences of the primers (FP-forward primer,RP-reberse primer) employed in the PCR reaction were:
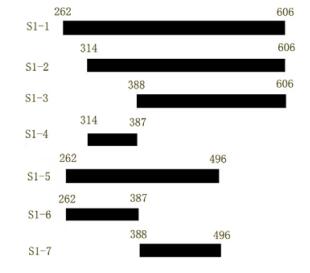
Figure 1. Schematic description of SARS-CoV S protein sequential deletion mutations. Numbers on the bars indicated the first or the last amino acid position in the deletion mutation. Mutant names are indicated in the left column.
S1-1 FP:5′-gtcagggatccatgctcaagtatgatgaaaatggtac-3′
RP:5′-actcgagaattcctaaacatcagtgcagttaac-3′
S1-2 FP:5′-ctgcagggatccaccatggtgagattccctaatattac-3′
RP:5′actcgagaattcctaaacatcagtgcagttaac-3′
S1-3 FP:5′-ctgcagggatccaccatggtcaagggagatgatg-3′
RP:5′-actcgagaattcctaaacatcagtgcagttaac-3′
S1-4 FP:5′-ctgcagggatccaccatggtgagattccctaatattac-3′
RP:5′-actcgagaattcctatacaaaagaatctgc-3′
S1-5 FP:5′-gtcagggatccatgctcaagtatgatgaaaatggtac-3′
RP:5′-actcgagaattcctaaactctgtaaggttgg-3′
S1-6 FP:5′-gtcagggatccatgctcaagtatgatgaaaatggtac-3′
RP:5′-actcgagaattcctatacaaaagaatctgc-3′
S1-7 FP:5′-ctgcagggatccaccatggtcaagggagatgatg-3′
RP:5′-actcgagaattcctaaactctgtaaggttgg-3′
These forward primers carried a BamHⅠ restriction site and reverse primers carried an EcoRⅠ restriction site. The amplified products were gel purified and cloned into the pET His vector. The ligation products were transformed into DH5 competent cells. The positive clones were identified by restriction analysis and sequencing.
-
Each recombined strain was grown in 2×YT medium and expression was induced by 1mmol/L isopropyl-β-D-thiogalactopyranoside (IPTG) at 37℃. Cells were harvested by centrifugation and the pellet was sus-pended in binding buffer(20mmol/L Tris HCl pH8.0,500mmol/L NaCl,5mmol/L Imidazole,1mmol/L NaF,and 1mmol/L PMSF),sonicated and centrifuged at 12 000g at 4℃ for 30 min. The pellet was resus-pended in urea buffer (8mol/L Urea,20mmol/L Tris HCl,pH8.0,500mmol/L NaCl,5mmol/L Imidazole,1 mmol/L NaF,1 mmol/L PMSF,and 1 mmol/L DTT),then the suspension was centrifuged at 12 000g at 4℃ for 30 min,and the supernatant was applied to Ni-NTA affinity column(Qiagen) equilibrated in urea buffer. The Ni-NTA affinity column was then washed with gradient washing buffer (8mol/L Urea,20mmol/L Tris-HCl,pH8.0,500mmol/L NaCl,5,20,40,80 and 100mmol/L Imidazole). Purified protein was eluted with elution buffer(8mol/L Urea,20mmol/L Tris-HCl,pH8.0,500 mmol/L NaCl,and 200 mmol/L Imidazole) and dialyzed with dialytic buffer(20mmol/L Tris-HCl,pH8.0,500 mmol/L NaCl,0.1% Triton X-100,and 0.2% BSA). Purified S1-1~S1-7 and ACE2 proteins were obtained and checked by SDS-PAGE.Only freshly purified proteins were used for further experiments.
-
S1-1~S1-7 proteins were separated by SDS PAGE in a 15% polyacrylamide gel and were transferred to nitrocellulose membrane,as described by Sambrook and Rus-sell. After blocking with PBST(14mmol/L NaCl,2.7mmol/L KCl,10mmol/L NaHPO4,1.8 mmol/L KH2PO4 and 0.02% Tween 20) containing 5% nonfat milk for 1h at 37℃,the membrane was incubated for 2h at 37℃ with human SARS positive sera(1:20 dilution). After washing three times in an appropriate volume of PBST,the membrane was incubated for 1h with horse-radish peroxidase (HRP) labeled goat anti human IgG (1:5 000,Sigma). The membrane was washed three times with PBST and once with distilled water,for 10 min each. Reagents for enhanced chemiluminescence were applied to the blots and the light signals were detected by X-ray film.ACE2 protein was separated by SDS-PAGE using a 12% polyacrylamide gel and was transferred to nitrocellulose membrane as described above. The membrane was incubated for 2h at 37℃ with the rabbit anti-serum against ACE2:Anti-veroACE2 (1:1000 dilution).
-
0.1 mg each of seven purified S proteins were mixed well with 1% Al(OH)3 respectively,and used to immunized rabbit by injecting hypodermal at three point. Three weeks after first immunization. Booster shot were performed,10 days later,the blood samples were collected and titers of antibodies were measured by ELISA.
-
6.1 Purified S1-1~S1-7 proteins,dissolved respec-tively in coating buffer (0.016mol/L Na2CO3,0.034 mol/L NaHCO3 pH9.6),were individually coated to a 96-well microplate(Nunc,Roskilde,Denmark) and incubated at 4℃ overnight. Wells were then blocked with 4% non-fat milk,0.5% BSA in phosphate buffer saline (PBS),incubated for 1h at 37℃,then washed with PBS and dried at 37℃ for 1h. The purified ACE2 protein(10 μg/mL) was then added to react with the S1-1~S1-7 proteins for 1h. Each well was washed 5 times with washing buffer (0.5% Tween-20 in PBS). The wells coated with ACE2 alone were used as the positive control. The empty wells with ACE2 only were used as a negative control. Another negative control was wells coated with S1-1 without addition of ACE2. The rabbit anti-serum specific against the ACE2 was diluted in sample buffer (1%BSA in PBS,1:1 000),then incubated at 37℃ for 30 min. Each well was washed 5 times with washing buffer then incubated with peroxidase conjugated goat anti rabbit IgG antibody at 37℃ for 30 min. The wells were washed again with washing buffer. The peroxidase reaction was visualized by using O-phenylenediamine as a substrate,after 10 min at 37℃ or until the color was visualized. The reaction was stopped by adding 50 μL/well of 1NH2SO4. The absorbance was read at 490 nm. The samples were run in triplicate.
In the interaction disruption assay,rabbit anti-S1-7 serum was added into the well before adding ACE2,the remainder of the steps were the same as above.
1.1. Plasmids,strains and reagents
1.2. Construction of the recombined Plasmids
1.3. Expression and Purification of recombined protein
1.4. Western blot
1.5. Preparation of rabbit anti-S1-1~ S1-7 sera
1.6. Interaction of S proteins with ACE2
-
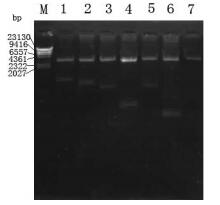
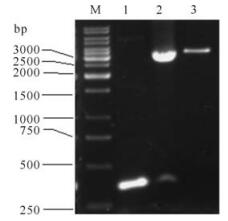
The fragments of 1 035bp (S1-1),879bp (S1-2),657bp (S1-3),222bp (S1-4),705bp (S1-5),128bp (S1-6) and 327bp (S1-7) were amplified by PCR using plasmid pGEM-S as template and were then cloned into prokaryotic expression vector pETHis. The corr-ected clones were confirmed by restriction analysis and sequencing. The results of agarose electrophoresis are shown in Fig. 2 and Fig. 3.
-
S1-1~S1-7 proteins were successfully expressed in E.coli BL21(DE3) at a high level and formed insoluble inclusion bodies. As Fig. 4 shows,the proteins were expressed and purified to a high purity.
-
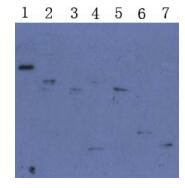
To investigate whether the induced proteins were the expected proteins,Western Blot analysis of purified proteins were processed. As Fig. 5 showed,all purified S1-1~S1-7 proteins could react with the SARS pat ients sera specifically and the molecular weights were consistent with expectations,indicating the approp-riate proteins were produced.
-
The recombinant ACE2 protein was successfully expressed in E.coli DH5. Fig. 6 showed that ACE2 was induced and purified. The purified ACE2 protein could react with anti-ACE2 serum and the expected 92kDa band was observed in Western Blot (Fig. 7).
-
Purified recombinant proteins S1-1~S1-7 were fixed in the 96-well plate by coating,and then purified ACE2 protein was added to allow their interaction. A well coated with purified ACE2 protein was used as positive control. Another coated with purified S1-1 protein without adding ACE2 protein was used as negative control. Then Anti-vero-ACE2 rabbit serum was added as first antibody and HRP labeled goat-anti-rabbit IgG was added as second antibody. ELISA showed that the results of S1-1,S1-2,S1-3,S1-5,S1-7 wells were positive,while the results of S1-4,S1-6 were negative (Table 1.). Our data showed that the aa 388~396 of S1 protein was essential to the interaction between S protein and ACE2.

Table 1. Interaction between truncated S protein and ACE2
To further confirm this result,an antibody induced by S1-7 which only contains aa 388~396 of S protein was used for disrupting the interaction between S and ACE2. The results show that all the interactions we re nearly completely blocked by this antibody (Table 2.). Whereas the antibody induced by S1-4 which contained deletions aa 388~396 did not show ability to block this interaction.

Table 2. Interaction blocking by antibody against S1-7
2.1. The construction of the recombined Plasmids
2.2. Expression and purification and recombinant proteins S1-1~S1-7
2.3. Western Blot analysis of purified proteins
2.4. The expression,purification and Western Blot analysis of recombinant ACE2 protein
2.5. The interactions between varies of recombi-nant truncated S proteins and ACE2 protein.
-
SARS coronavirus is a newly discovered etiologic agent of SARS. To date,SARS vaccine in clinical experiments is the wild-type inactivated virus (1). Ho-wever,this vaccine is potentially hazardous because of the risks of the virus becoming reactivated. Engineering a protein vaccine for SARS,therefore,is a relatively safer option and has great potential as a future direction in SARS vaccine development.
The spike protein of SARS-CoV is composed of two parts: S1 and S2. S1 is involved in viral infection,pathogenesis,host ranges and binding to the receptors (5, 6). So this study on the interaction between S1 protein and the receptor ACE2 may shed light on the development of engineering protein vaccines of SARS.
In this study,seven truncated S1 sequences (S1~S7) were cloned in the prokaryotic expression vectors and induced to express at a high level. Our results showed that S1-1,S1-2,S1-3,S1-5 and S1-7 can interact with purified ACE2 protein specifically,but S1-4 and S1-6 cannot interact with this receptor. S1-1,S1-2 and S1-3 are sequential truncated proteins of the N-terminal S protein. They can both bind to ACE2,suggesting aa 262~387 is not essential for S1-ACE2 interaction. S1-5 can bind to ACE2,while S1-6 can't and S1-7 can bind to ACE2 while S1 4 can t,both of which suggested that aa 388~496 is essential for the S1 ACE2 interaction.
In this study,two methods were used for investiga-ting the binding of various truncated S1 proteins to the ACE2. The first one was the Far-Western Blot. Various truncated S1 proteins were resolved on a 15% SDS-PAGE gel and then transferred onto N.C membrane. The blots were overlayed with ACE2 and the binding of ACE2 protein was detected by X-ray with anti-ACE2 rabbit serum as the first antibody and goat-anti-rabbit IgG as the second antibody. However,no ACE2 protein binding to the membrane was detected. The second method was performed as follows. Truncated S1 proteins were fixed onto the plates well by coating without denaturing,and then purified ACE2 protein solution was added to allow the interaction to proceed.Anti-ACE2 rabbit serum was used as the first antibody and goat-anti-rabbit IgG was used as the second antibody. HRP activities were assayed and our results showed that S1-1,S1-2,S1-3,S1-5,S1-7 can all bind to purified ACE2 protein specifically,while S1-4 and S1-6 can not interact with ACE2. The results in these two experiments are different suggesting that denaturing of various truncated S1 proteins in SDS-PAGE in the first experiment disrupt the conformation of S1 proteins. But in the second experiments,all these proteins maintained their activated conformation. Our data indicated that the interaction of S1 protein and receptor ACE2 perhaps depend on the conformation of S1 protein.
Our data showed that anti-S1-7 protein rabbit antiserum could block the interactions between ACE2 and truncated S1 proteins (S1-1,S1-2,S1-3,S1-5,S1-7). So the recombinant S1-7 protein could induce protective neutralizing antibody against SARS-CoV. Our study sheds light on the development of engineering protein vaccines for SARS.














 DownLoad:
DownLoad: