-
Investigation of the expression and replication of the HBV genome as well as the full viral life cycle has been hampered by the lack of an in vitro tissue culture system in which HBV is propagated. The HepG2.2. 15 cell line is a putative HBV cellular model and it have been used for antiviral research for many years (9, 3, 7) ,however,HBV replication in HepG2.2.15 cells is very different from HBV natural replication in vivo. HBV covalently closed circular DNA(cccDNA),a key viral replicative intermediates in viral life cycle,is absent in this cell line. Here we constructed a HBV replicon containing a 1.3× copy of the HBV genome,and established a transient transfection cellular model of HBV replication.
HTML
-
Restriction endonuclease AatⅡ,NsiⅠ,BglⅡ and BclⅠwere purchased from NEB Co.,Pfu DNA polymerase,T4 DNA ligase were purchased from Promega Co.,Minimal essential medium(MEM),Fetal bovine serum(FBS) and lipofectamine were purchased from Invitrogen Co.
-
TA cloning vector pCR2.1,competent cells TOP10 and INV110 were purchased from Invitrogen Co. Plasmid pHBV-dimer,which harbors a head-to-tail dimer of the HBV(ayw) genome,was kindly provided by Prof. Mengji Lu(Institute of Virology,Duisburg-Essen University).
-
A human hepatoblastoma cell line,Huh7,were maintained in MEM medium supplement with 10% fetal bovine serum(FBS) at 37℃ in a moist atmosphere containing 5% CO2 in air.
-
Primers P1 and P2 were designed based on HBV gene sequence(GenBank accession number:AB042282),P1(1070-1088):5'-GTATACAAGCTAAGCAGGC-3' andP2(1984-1964):5'-TCGAATAGAAGGAAAGAAG-TC-3'for amplifying HBV gene from nt 1070 to nt 1984.
-
A HBV replicon was constructed from pHBV-dimer. Firstly,a 915bp fragment (nt 1 070~1 984) of the HBV gene was amplified from pHBV-dimer,and cloned into pCR2.1.A plasmid containing a forward HBV gene fragment was identified and named as pHBV0.3. Secondly,a 2 852bp fragment of the HBV genome was digested from pHBV-dimer with AatⅡand NsiⅠ,and ligated with pHBV0.3,which was digested with AatⅡand Nsi Ⅰ; the recombinant plasmid was identified and named as pHBV1.0. Finally,a 921bp fragment of the HBV genome was digested from pHBV-dimer with NsiⅠand Bgl Ⅱ,and ligated with pHBV1.0,which was digested with NsiⅠand BclⅠ,the recombinant plasmid was identified and named as pHBV1.3.
-
Huh7 cells were seeded at 5×105 cells/well in 6-well plates.24 h later,5μg pHBV1.3 plasmids were transfected with 5μL of lipofectamine reagent according to the manufacturer's instructions. Fresh media were added and maintained for 96h. HBsAg,HBeAg,HBV DNA,and HBV replicative intermediates and transcripts were measured everyday.
-
Huh7 cells were transfected with pHBV1.3 as described above; cells were passaged at 4 day intervals. Intracellular HBV replicative intermediates were detected at day 4,8,12 and 16.
-
HBsAg,HBeAg in supernatant were detected by ELISA according to the manufacturer's instructions. HBV DNA was quantified by real-time PCR. In brief,200 μL supernatant were treated with 50 μg /mL of DNase Ⅰfor 30min at 37℃,and total DNA was extracted and measured by real-time PCR. Intracellular HBV repli-cative intermediates and transcripts were mea-sured by Southern blot and Northern blot respectively. In brief,total cellular DNA and RNA were isolated,5 μg of total DNA and 20μg of RNA were loaded on a 1.5% agarose gel and formaldehyde agarose gel respectively,and transferred to a nylon membrane,then hybridized with a 32P-labeled HBV DNA probe and analyzed with storage phosphor system (Cyclone,Packard Co.)
-
Huh7 cells were transfected with pHBV1.3 as described above. Fresh medium containing 0.01,0.1,1 and 10 μmol/L adefovir (Gilead Sciences,USA) were added 24h later,and maintained for 72 h. Total cellular DNA were isolated,intracellular HBV replicative intermediates were measured by Southern blot as described above.
1.1. Reagents
1.2. Plasmids and bacteria
1.3. Cells culture
1.4. Primers design
1.5. Construction of HBV replicon
1.6. Transient transfection of pHBV1.3
1.7. Persistence of HBV replicon
1.8. HBV replication and expression
1.9. Antiviral test in Huh7 cells transfected with pHBV1.3
-
A 915 bp fragment was obtained by PCR. This fragment was inserted to pCR2.1 vector. Plasmids contai-ning forward fragment were identified by restriction endonuclease analysis and named as pHBV0.3,sequence analysis showed that there was no mutation in the inserted fragment. A 2852 bp fragment was obtained from pHBV-dimer by digestion with AatⅡ and NsiⅠ,and inserted into pHBV0.3,plasmids containing 1.0 fold of HBV genome was constructed and named as pHBV1.0. A 921 bp fragment was obtained from pHBV-dimer by digestion with NsiⅠand BglⅡ,and inserted into pHBV1.0; plasmid pHBV1.3 was identified by restriction endonuclease analysis. After digestion with AatⅡ,There were two fragments of 3 200 bp and 3 856 bp (Fig. 1).
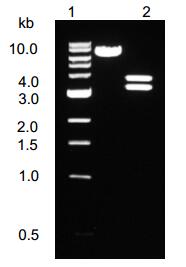
Figure 1. Restriction endonuclease analysis of plasmid pHBV1.3. pHBV1.3 was digested with restriction endonuclease AatⅡ,the cleaved fragments were separate by electrophoresis in a 1% agarose gel,The feagments are 3 200 bp and 3 856 bp (lane3). Plasmid pHBV1.3 is 7 056 bp (lane2). The molecular weight standard is 1 kb DNA ladder (lane1).
-
Plasmids were transfected into Huh7 cells and allowed to replicate for 96 h.Quantities of HBsAg and HBeAg present in the supernatant were measured by ELISA; quantity of HBV DNA in supernatant was determined by real-time PCR; intracellular replicative intermediates and transcripts were analyzed by Southern blot and Northern blot respectively. The levels of HBsAg and HBeAg reached a plateau within 72 h. Further incubation did not increase the concentration of HBsAg and HBeAg in the culture supernatant. There was a well consistency of the levels between HBsAg and HBeAg in supernatants (Table 1.). Meanwhile,the levels of HBVDNA in the culture supernatant were 6.5×102copies/mL,1.2×103 copies/mL,2.5×105 copies/mL and 2.7×105 copies/mL at 24 h,48 h,72 h,and 96 h respectively. Furthermore,the intensity of the hybridi-zation signal,for intracellular replicative intermediates and int-racellular transcripts also reached a maximum within 72 h (Fig. 2). The results indicated that the HBV replicon could effectively initiate HBV replication and expression of HBV proteins.

Table 1. HBV products in culture supernatant after transfection with HBV replicon(x±s)
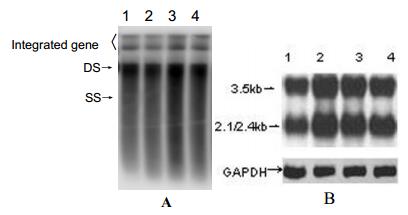
Figure 2. Intracellular HBV replicative intermediates (A) and intracellular HBV transcripts RNA (B) in Huh7 cells transfected with a HBV replicom. A: Southern blot analysis of 5μg of total DNA extracted from Huh7 cells transfected with pHBV1.3. and probed by a 32P-labeled HBV-specific DNA probe. Bands corresponding to the expected size of double-stranded (DS) and single-stranded (SS) HBV DNAs are indicated. B: Northern blot analysis of 20μg of total RNA was extracted from transfected cells. and hybridized with 32P-labeled HBV-and GAPDH-specific probe. Bands corresponding to the expected size of 3.5kb and 2.1/2.4kb HBV transcripts are indicated. Lane1-4 : 24h,48h,72h and 96h. post transfectionre spectively.
-
Huh7 cells transfected with pHBV1.3 were passaged at an interval of 4 days. Southern blot analysis shown that intracellular HBV DNA replicative intermediates could be found for at least twelve days. The results indicate that the HBV replicon can persist for an extended time,even after cell division.
-
A cell-based antiviral assay was used to test the adefovir activity. Huh7 cells were transfected with pHBV1.3,and treated with different concentrations of adefovir on the next day. Southern blot analysis indicated 10 mol/L of adefovir could completely inhibit HBV replication in Huh7 cells transfected with plasmid pHBV1.3,and the inhibition is dose-dependent (Fig. 3).
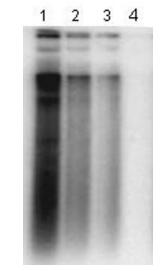
Figure 3. Antiviral test of in vitro system transfected with HBV replicon. Huh7 cells were transfected with plasmid pHBV1.3.and then treated with 0.01,0.1,1 and 10μmol/L of adefovir for 72 h. At the end of the treatment,total DNA was isolated and analyzed by Southern blot. Lane1-lane4: total DNA from Huh7 cells treated with 0.01,0.1,1 and 10μmol/L of adefovir respectively.
2.1. Construction of plasmid pHBV1.3
2.2. Characterization of HBV replicon in vitro
2.3. Persistence of HBV replicon in Huh7 cells
2.4. Adefovir activity analysis in Huh7 transfected with HBV replicon
-
The hepatitis B virus(HBV) contains a 3.2-kb,circular,double-standed DNA genome and causes acute and chronic hepatitis,cirrhosis,and hepatocellular carcinoma (5, 10). Because of the limited host range of HBV and the inability to grow it in vitro,much of what is known about the HBV life cycle has come from studies of other members of the hepadnaviridae family,especially the woodchuck hepatitis virus(WHV) and the duck hepatitis B virus(DHBV),which infect their native species and,in the case of the DHBV,can be grown in tissue culture (1, 8, 11).
The HBV cellular model system has been the subject of many studies in the last four decades,unfortunately,so far no group has reported the identification of an appropriate tissue culture system to propagate HBV. This has greatly hampered the development of HBV studies in many fields,such as virology,molecular biology,immunology and antiviral therapy. In an attempt to overcome this obstacle,several groups transferred the HBV genome into human hepatocytes via plasmids,in which the HBV gene could replicate,express,and even assemble infectious virions (9, 12). Because the HBV genome is very compact and all genes are over-lapped with each other,HBV genome were inserted into plasmids in a head-to-hail linkage manner. Sells et al(1987) transfected plasmids containing two head-to-tail copies of the HBV genome into the heptoblastoma cell line HepG2 to obtain the HepG2.2.15 cell line. This popular cell line has been widely used in antiviral studies as a putative HBV cellular model.However,HBV replication in this cellular system is very different from natural HBV replication in vivo because the HBV genome is integrated with a cellular chromosome and the covalently closed circular DNA(cccDNA) is absent. Its use is therefore limited to antiviral and replication studies of HBV.
Recently some group have transfected an overlength HBV genome into hepatocytes for HBV replication and anti-HBV studies. It is well known that the longest HBV transcript is a 3.5 kb mRNA; therefore,several plasmids carrying multiple HBV genomes were constructed,such as 2.0 copies,1.3 copies,1.2 copies and 1.1 copies. The 1.3-fold HBV genome contains complete HBV repli-cation units (such as EnhⅠ, EnhⅡ,DR1,DR2,the transcription origin site of the viral pre-genome,promoter and ORFs),and its replication efficiency is markedly higher than that of others (2, 13) ,therefore,a plasmid vector containing a 1.3-fold HBV genome was preferen-tially considered. We constructed a HBV replicon based on the plasmid pHBV-dimer which carried two head-to-tail copies of the HBV genome. The HBV 1.3-fold genome was cloned into the pCR2.1 vector by combined PCR and restriction endonuclease digestion. The same enzyme sites for the HBV genome and plasmid pCR 2.1 vector were used,such as NsiⅠ and BglⅡ/BclⅠ. The restriction endonuclease BglⅡand BclⅠ used for inserting the 921 bp fragment into pHBV1.0 have similar function,BglⅡ recognition sequence is A↓GATCT while BclⅠrecognition sequence is T↓GATCA (arrow shown cleavage site),so DNA sequence from pHBV-dimer with BglⅡdigestion could be inserted to linear pHBV1.0 that digested with BclⅠdigestion. But BclⅠwas sensitive to dam methylase,and general bacteria but not INV 110 possess dam methylase,with which the sequence GATC will be methylate sequence GmATC,so INV 110 were selected to serve as competent cells.
After HBV replicon was transfected into Huh7 cells,HBV genome could effectively replicate and express. There were high levels of HBsAg,HbeAg and HBV DNA in cultures supernatants and high levels of intra-cellular HBV replicative intermediates and intracellular HBV transcripts. Comparing with HBV replication in HepG2.2.15 cell line,it was much more similar to natural status of HBV infection. Furthermore,HBV replicon was able to replicate in many kind of hepatocytes,such as Huh7 cells,HepG2 cells (data not shown) and primary human liver cells. Therefore,this cellular system has much more advantage in study of antiviral agents and viral replication than that of HBV genome integrated hepatocyte system. The intracellular HBV replicative intermediates could be probed after transfected cells were passaged for three times (12 days),this result indicated HBV replicon could persist for a long time even after cell division. But HBV genome will be progressively lost because of continuous cell division. Furthermore,adefovir,a novel antiviral drug (6, 4) ,reduced replication of HBV in this cellular model,it was shown that this cellular model system might be used for study of anti-HBV agents in future.














 DownLoad:
DownLoad: