-
For most viruses the replication cycle is characterised by a temporal cascade of virus-induced gene expres-sion. Immediate and delayed early genes alter the cellular environment and produce enzymes that allow the infected cell to replicate the viral genome. Late genes are expressed after viral genome replication and encode the structural proteins required for production of novel virion particles that spread infection to new host cells. Baculoviruses deviate from this pattern by encoding an additional very-late class of genes. This very-late phase of protein expression corresponds to a shut down of virus budding (and budded virion (BV) production), a retention of nucleocapsids in the nucleus and occlusion of these nucleocapsids into paracrystalline protein occlusion bodies (OB, also sometimes referred to as polyhedral inclusion/occlusion bodies).
The p10 gene is one of two baculovirus genes hyper-expressed very late [10-15 hours post-infection (hpi) onwards] during infection (50-52). The role of the other hyper-expressed very-late protein, polyhedrin (Polh), as the main component of the OB, in the protection of virions from environmental damage and in the horizontal transmission of virus between hosts, has been extensively characterised and is very well understood (20).
After over three decades of research, however, the role of the p10 gene product remains a mystery. Most of the early research on P10 has been extensively reviewed by van Oers and Vlak (58) and we do not intend to reiterate their detailed description of early P10 work, particularly regarding analysis of p10 promoter activity and transcription patterns. In this review we take a look back at some of the earlier studies relating to the function of the p10 gene product and attempt to reinterpret these results based on more recent work over the past decade. We also explore whether a model for P10 function and its role in infection can be inferred from combining our experi-mental knowledge with an analysis of P10 sequence data taken from the ever increasing database of fully sequenced baculovirus genomes.
HTML
-
The P10 protein gets its name from the predicted mass of the protein expressed from the Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) p10 gene (33). Physical analysis of the expressed protein by SDS-PAGE, however, estimated its size at 7.2 kDa (1), 7.5 kDa (60) and 8 kDa (50). It is generally accepted that expression of the P10 protein is non-essential for baculovirus infection, replication and spread and that disruption of the p10 open reading frame (orf) does not have a detrimental effect on infection either in cell culture or in a caterpillar host (15, 59).
-
Between the 1960s and 1980s a number of transmission electron microscopy (TEM) ultrastructural studies of baculovirus-infected cells were published, describing the intracellular and intranuclear changes that take place during infection. These studies went a long way to describing the process of virion occlusion into the large paracrystaline OBs. Summers and Arnott (54) were the first to describe the formation of a second distinct infection-associated structure in the nucleus and cytoplasm of Trichoplusia ni multicapsid nucleopolyhedrovirus (TnMNPV), -infected Trichoplusia ni (T. ni) caterpillars midgut cells, consisting of what they referred to as fibrous material. Similar structures were also observed in in vitro cultured T. ni cells infected with TnMNPV (41) and in vitro cultured Spodoptera frugiperda cells infected with S. frugiperda nucleopolyhedrovirus (SfNPV) (32) or AcMNPV (11). Examples of these structures in both nucleus and cytoplasm of infected cells is shown in figure 1A. These structures appeared to consist of parallel bundles of fibrous material (shown in a close up view in figure 1B) described as possessing a longitudinal periodicity of about 70 Å (54) and forming a macromolecular lattice pattern (14). These fibrous bundles showed an intimate association with OBs (Fig. 1C) at various stages of formation and with electron dense structures (Fig. 1D) called electron dense spacers (ES) (41), "membrane-like profiles" (54), or "cisternae" (26). In both associations the fibrous bundles were oriented perpendicularly to the surface of OBs and ESs.
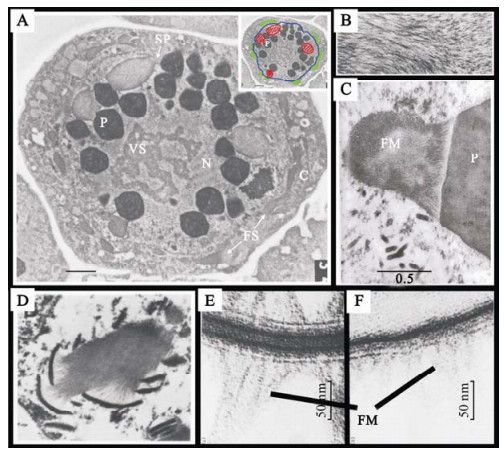
Figure 1. Transmission electron microscopy ultrastructural studies of P10 associated structures. Ultrastructural studies of nucleopolyhedrovirus infected cells revealed Fibrous material forming extensive structures in the nucleus and the cytoplasm. Panel A (taken from ref. 59) shows a transmission electron micrograph of a Spodoptera frugiperda cell infected with AcMNPV 48 hpi. FS, Fibrous structure; N, nucleus; C, cytoplasm; P, polyhedral occlusion body; VS, virogenic stroma; SP, electron dense spacer. The insert shows the localisation of the nuclear envelope (blue) and the location of nuclear (red) and cytoplasmic (green) fibrous structures. Panel B (taken from ref. 11) shows a close up view of fibrous structures in AcMNPV infected Spodoptera frugiperda cells showing the lattice mesh structure of the fibres. Panel C (taken from ref. 54) shows a bundle of fibrous material (FM) along side a polyhedral occlusion body (P). Fibres can be seen intimately interacting with the surface of the occlusion body, lying perpendicular to its surface. Small fibres can also bee seen projecting from other facets of the OB. Panel D (taken from ref. 32) shows the intimate association of electron dense spacer structures with fibrous material. The interaction of fibrous material with ES structures is similar to the interaction seen with OBs. Panels E and F (taken from ref. 11) are a side by side structural comparison of electron dense spacer structures (E) and the polyhedral envelope (F) showing the similarity between the two. Fibrous material (indicated by FM) can be seen projecting away from both of these structures.
Detailed TEM analyses of ES structures and the surface of OBs revealed that the envelope that surrounds mature OBs was almost identical in structure to the ES membranes (See Fig. 1E and 1F) (11, 26). There is some evidence suggesting fibrous material may condense at the OB periphery to form the polyhedral envelope (11). Whether ES structures are precursors of the polyhedral envelope or represent condensation of extraneous surplus fibrous material remains unclear.
Immunogold-labelling TEM studies indicated that P10 showed an association with fibrous structures (55), which was absent when P10 function was 'knocked-out' by disruption of the p10 orf (15, 59, 63). P10 has also been reported to associate with the ES and envelope structures surrounding the OBs (35, 59). P10 association with OBs has also been observed by biochemical fractionation (59) but was often dismissed as a contaminant. A high level association of P10 with the surface of mature OBs upon their release from infected cells has been documented by immuno-fluorescence confocal microscopy (6).
The role of P10 in the formation of the polyhedral envelope remains unclear. In one study, in which LacZ was fused to the amino-terminal 51 amino acids of P10, ES structures failed to form (59). Instead this fusion protein formed amorphous aggregates in the nucleus of infected cells. There are conflicting reports as to whether or not this mutant virus formed an intact polyhedral envelope (58, 59)). Separate and independent studies utilising similar strategies to knock-out P10 function, either by lacZ/P10 fusion (63) or phospho transferase/P10 fusion (15), found that both ES structures and polyhedral envelope formed normally. The polyhedra of these mutant constructs did appear to be more fragile and possess a more loosely associated envelope (63).
P10 from Orgyia psuedotsugata multiple nucleopoly-hedrovirus (OpMNPV) and from the closely related AcMNPV have both been described as forming cytop-lasmic filament-like structures from about 18 hpi onwards, which mature into a continuous network of thick rod-shaped or tubule structures (shown in detail in Fig. 2) from 30-36 hpi onwards (6, 48). The P10 filaments show a strong association with thickened bundles of cytoplasmic microtubules (Shown in yellow in Fig. 2) (6, 61). Similar filament and tubule structures have been reported to be formed by the filament-associated late protein of entomopoxviruses (FALPE) but FALPE filaments did not show an association with host cell microtubules (2). Formation of P10 filaments does not require the expression of other baculovirus proteins (3, 17, 18) but requires intact host-cell microtubules (6, 46). P10 interaction with microtubules has been biochemically confirmed by yeast-2-hybrid and co-immunoprecipitation studies (46). P10-independent microtubule reorganisation occurs early in baculovirus infected cells, and eventually the microtubule network breaks down completely (61). P10 is required and sufficient for the late phase microtubule reorganisation into thick cables in infected cells and shows similar microtubule reorganisation properties in cells transiently expressing P10 (6).
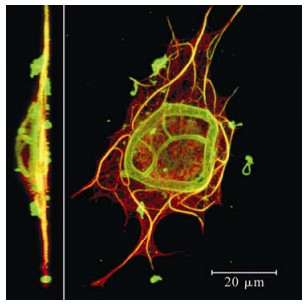
Figure 2. Immunofluorescence confocal microscopy 3D recons-truction analysis of P10 structures. Side on (insert) and top down view of an AcMNPV infected TN-368 cell 48 hpi stained for P10 (green) and tubulin (red) (taken from ref. 6). P10 filaments (which form in the cytoplasm from 18 hpi onwards) show a strong association with thickened bundles of microtu-bules (shown in yellow where the green and red signal colocalise). P10 tubules (which form perinuclearly from 30 hpi onwards) form a dome shaped cage around the nucleus (seen clearly in the side on view). Scale bar indicates 20 mm.
Historically P10 research has focused on the potential role of the nuclear P10 FBs. Because of the very-late phase expression of P10 and the intimate association of P10-associated structures with OBs, it is easy to assume that P10 is intimately involved in the nuclear processes of OB formation and virus occlusion.
Some of the earliest ultrastructural studies describe cytoplasmic FB structures, which displayed slightly different packing and electron density properties and possessed a smaller cross sectional profile to nuclear FBs (54). Recent data suggest that cytoplasmic FBs show a higher level of P10 association than nuclear FBs (6). These cytoplasmic FBs possess a tubule-like staining pattern and form a complex perinuclear cage, sometimes seen to lie adjacent to a network of perinuclear microtubules (6, 54). Cytoplasmic FBs have also been described as associating with vesicles and ribosomes (54). Many of the cytoplasmic FB structures previously described by TEM probably represent cross-sections of the tubules, described by immunofluorescence, that project over the nucleus (Compare figure 1A insert and the 3D organisation of the perinuclear cage forming a dome over the cell shown in Fig. 2) It is often difficult to identify nuclear FBs by immunofluorescence without extensive digital enhancement of micrograph datasets, due to the low level of staining by anti-P10 antiserum, however, P10 association with both nuclear FBs and maturing OBs has been reported (6). It is clear from both past ultras-tructural studies and from these immunofluorescence studies that the nuclear FBs (sometimes called nuclear worms due to their distinctive staining pattern) display different structural properties compared to the cytoplasmic perinuclear FB tubules (Fig. 3).
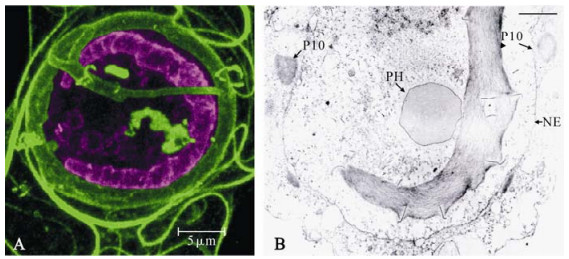
Figure 3. Nuclear P10 structures differ in structure from cytoplasmic P10 structures. Panel A: Enhanced view of the P10 staining signal in the nuclear region of an AcMNPV infected TN-368 cell 48 hpi imaged by confocal laser scanning microscopy. The signal corresponding to the intranuclear region was artificially enhanced and coloured purple (using Adobe Photoshop) to distinguish the intranuclear structures from the cytoplasmic structures coloured green. The signal enhancement revealed the presence of P10 associated OBs and revealed the irregular shape and staining pattern of the nuclear P10 tubule, often referred to as a P10 "worm". This image emphasises the structural differences between the nuclear and cytoplasmic (perinuclear) P10 associated structures. Scale bar indicates 5 mm. Panel B: Transmission electron micrograph of AcMNPV infected Spodoptera frugiperda cell at 48 hpi provided by A Patmanidi (Oxford Brookes University) showing electron dense spacer structures extruding from ridges in the P10 tubule (labeled P10). A polyhedral occlusion body has been labelled PH and the nuclear envelope is labelled NE. These ridges may correspond to those seen along the "worm" structure shown in panel A.
-
The past decade has seen an explosion in the number of published baculovirus genomes. The National Centre for Biotechnology Information (NCBI) Viral Genome Project reference sequence database (http:// www.ncbi.nlm.nih.gov/sites/entrez) currently contains 27 Alphabaculovirus (Lepidopteran nucleopolyhed-roviruses (NPVs)) genomes, 8 Betabaculovirus (Lepi-dopteran granuloviruses (GVs)) genomes, 3 Gamma-baculovirus (Hymenopteran NPVs) genomes and 1 Deltabaculovirus (Dipteran NPV) genome. P10 homo-logues have been identified in all of the sequenced Alphabaculovirus genomes and in 2 of the sequenced betabaculovirus genomes. No P10 homologues have been identified in either the Gammabaculoviruses or the Deltabaculoviruses. The Entomopox P10 homo-logue, FALPE, identified in the genome of Amsacta moorei entomopoxvirus (AmEPV), a lepidopteran virus from the Poxviridae virus family (2), shares structural and functional features with baculovirus P10 (3).
Previous bioinformatic analysis of 9 different bacu-lovirus P10 gene sequences (58) revealed that bacu-lovirus P10 homologues share a relatively low level of sequence homology when compared to the polh gene. For example, OpMNPV and AcMNPV Polh and P10 show 90% (65) and 41.8% (58) sequence identity respectively at the amino acid level. All P10 proteins were found to share a structural and organisational homology. Van Oers and Vlak divided the P10 protein up into three distinct regions based on amino acid composition and phenotypic analysis of carboxy terminal deletion mutants (6, 58). These included; an amino terminal coiled-coil domain (consisting of over two thirds of the protein sequence); a proline rich region; and a carboxy terminal basic (lysine, arginine and serine rich) region. Additionally they described a region of variable length and sequence linking the proline rich and basic regions. A recently published bioinformatics study that included 30 different P10 homologues (including most of the ones identified in the sequenced genomes and several additional sequences available in the NCBI sequence database) found that this structural conservation held true across all bacu-lovirus P10 homologues (6). Based on this study we propose a slight alteration in the description of P10 structural organisation. The proline rich domain, the variable domain and the carboxy-terminal basic domain are each too small to form separate structural domains. Instead P10 likely adopts a lollipop-like structure consisting of a stalk (the coiled-coil domain) and a globular domain possessing a proline rich region and a basic region (Fig. 4). The sequences corresponding to the globular domain show very low levels of similarity between homologues that are not closely related. We therefore define this entire domain as the variable domain. In most P10 homologues this domain consists solely of a proline region, containing predominantly negatively charged amino acids, followed by a carboxy terminal positively charged basic region. It is only the sequences closely related to AcMNPV that possess additional regions of positive charge and negative charge between the proline rich and carboxy-terminal regions (Fig. 5).

Figure 4. Structural domains of P10. The structural organisation of P10 homologues is highly conserved. P10 consists of two structural domains; a coiled-coil domain with highly conserved amino-terminal 13-14 amino acids, and a variable domain containing a proline rich region and a positively charged basic region. In some P10 homologues these regions are separated by an additional basic and acidic region (see Fig. 5 below).

Figure 5. Alignment of baculovirus P10 homologues showing conserved structural organisation. Sequence alignments of type Ⅰ and type Ⅱ baculovirus P10 sequences using the ClustalX software package (described in ref. 6) manually shaded using the GeneDoc software package to highlight conserved structural features that characterise P10 homologues. Negatively charged acidic residues (aspartate (D) and glutamate (E); X) and positively charged basic residues (lysine (K), arginine (R) and histidine (H); X) were shaded to show the charge distribution within P10 homologues. Proline residues (P) were shaded to highlight the proline rich regions in P10 homologues. Glycine residues, known to disrupt α-helix stability, were also shaded (G). Small hydrophobic residues (usually valine (V), leucine (L) or isoleucine (I) and in some cases methionine (M) or alanine (A)) that occupy positions a and d of the abcdefg heptad repeat motif were shaded to highlight the position and register of the heptad motifs (X; positions that define a 3 aa element (abc) of the heptad repeat, X; positions that define a 4 aa element (defg) of the heptad repeat). Positions a and d occupied by other residues were also shaded (X and X respectively). Alignments were manually corrected by eye to better group the different regions.The shading clearly defines three separate regions; a large coiled-coil domain, a negatively charged proline rich region and an acidic region.
The high level of sequence similarity of the coiled-coil domain is concentrated within the amino-terminal 13-14 amino acids (6). P10 sequences can be divided into two distinct types (type Ⅰ and type Ⅱ, shown in Fig. 6) corresponding to a phylogenetic split previously described in the Alphabaculoviruses (25, 29) based on the sequence of these amino-terminal amino acids.
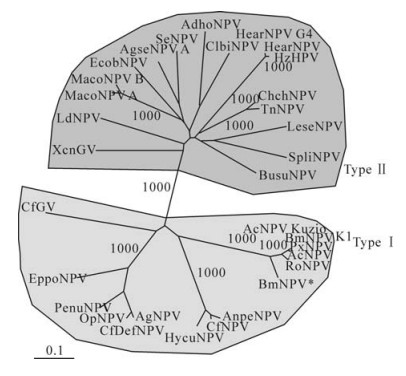
Figure 6. P10 phylogeny. Unrooted phylogenetic Neighbour-Joining tree generated from an alignment (calculated using the ClustalX software package) of 32 P10 sequences (described in ref. 6). Based on this tree P10 sequences have been defined as either type Ⅰ or type Ⅱ. Numbers indicate bootstrap values based on 1000 iterations. A number of interesting phylogenetic divisions showing high (1000) bootstrap values are indicated including the type Ⅰ and type Ⅱ split.
-
The P10 coiled-coil domain, which encompasses over two thirds of the entire protein, is defined by the characteristic heptad repeat motif (described below) containing between 7 and 11 heptad motifs (58). It has been suggested [based on recent studies of transiently expressed truncated P10 green fluorescent protein (GFP) fusion constructs] (17, 18) that the coiled-coil region of both Helicoverpa armigera nucleopoly-hedrovirus (HearNPV) and AcMNPV is essential and sufficient for P10 filament formation. AcMNPV P10 and HaEPV FALPE have both been shown by yeast-2-hybrid to self-associate, and this property has in both cases been directly mapped to the coiled-coil domain (3).
The structure and function of coiled-coils (39) is probably better understood than any other protein structural element. Coiled-coils consist of homo-or hetero-oligomeric bundles of α-helical protein chains, which can be arranged either parallel or anti-parallel. Coiled-coils can consist of two, three or four helices coiled around each other, although structures containing five or more coils have been reported. Coiled-coil domains are generally involved in the oligomerisation of multimeric proteins.
Normally α-helices have a periodicity of about 3.6 residues per turn (10). When α-helices coil around each other at a 20˚ angle they become slightly distorted, changing the periodicity to 3.5 residues per turn or 7 residues every 2 turns with respect to the supercoil axis. The defining property of coiled-coil domains is the organisation of its amino acid side chains in such a way to allow a "knobs-into-holes" interaction, where a residue from one helix fits into the free space sur-rounded by four residues from the facing helix (13, 14). This interaction is achieved by the incorporation of small hydrophobic residues (such as leucine, iso-leucine and valine and to a lesser extent methionine and alanine) at the 1st and 4th position of each 7 amino acid stretch (12, 40), usually referred to as positions a and d of the abcdefg heptad motif (Fig. 7). When mapped onto a model of an α-helix these positions line up on one side, forming a strongly amphipathic helix, in such a way that a hydrophobic region down one side forms the interface between two or more neigh-bouring helices. The specificity (homo-or heteromeric), orientation (parallel or anti-parallel) and magnitude (dimer, trimer or tetramer) is determined by the structure of the hydrophobic region and by complementary charges on adjacent helices at the e and g positions.
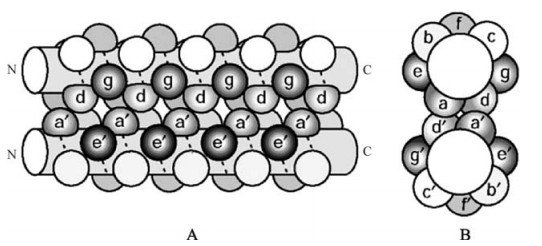
Figure 7. The coiled-coil heptad motif amino acid residue distribution. (A) side view and (B) top down view of a schematic representation of the amino acid residue localisation within a parallel two stranded α-helical coiled coil. The positions of the heptad repeat motif residues are indicated (abcdefg for one helix and a'b'c'd'e'f'g' for the other helix). The residues at positions a and d form the hydrophobic core of the helix-helix interaction. Interactions between residues at positions e and g help stabilise the interaction and contribute to the determination of the oligomerisation state and orientation of the interaction. (Taken from ref. 19).
This recognisable heptad repeat pattern makes it easy to identify potential coiled-coil regions from sequence data using mathematical techniques. A number of software packages are freely available on the web that calculate the probability of a region being a coiled-coil (24), one of which, COILS (40), has been used by Wilson et al. (64) to confirm the presence of coiled coil domains in a number of baculovirus P10 homologues. type Ⅰ sequences possess 7 heptad motifs while type Ⅱ sequences can have 8, 9 or 10 repeats. All P10 sequences, except the CfGV sequence, possess a 3 amino acid discontinuity of the heptad repeat pattern known as a "stammer". This stammer is an additional 3 residue element that corresponds to a repeat of the abc residues of a heptad motif (5). The change in position of the hydrophobic seam on the α-helix caused by the heptad motif phase shift induces additional over-winding of the coiled-coil structure. This stammer may be present to compensate for the presence of a proline residue (which preferentially forms a left-handed relaxed helix conformation rather than the right handed α-helix of the heptad repeat) and the helix breaking glycine residues (a non-chiral amino acid that due to the lack of a bulky side-chain can adopt a broader range of orientations). Alter-natively the proline and glycine residues may be present to stabilise the slight conformational shift caused by the stammer. The presence of proline in an α-helix has been shown to introduce a 30˚±10˚ bend in the struc-ture of the helix (8). It is unknown whether the com-bination of the proline and the stammer will stabilise or destabilise the coiled-coil interaction of P10. The presence of glycine residues near to this bend region in many P10 sequences suggests a requirement for additional flexibility. The biological significance of the coiled-coil distortion has not been determined, however, Dong et al. (17) found that deletion of the stammer element (thus bringing the heptad motif in frame along the entire helix) did not prevent AcMNPV P10 from aggregating into filamentous structures.
The higher order organisation of the coiled-coil is dependent on the properties of the amino acids at positions a, d, e and g as these are the residues that determine the chemical properties of coil interacting region. The properties of these residues is highly conserved among P10 sequences, particularly at positions a and d. The residues at positions e and g are conserved among closely related sequences. The current understanding of the determinants of higher order coiled-coil organisation do not yet reliably allow us to predict the oligomerisation state directly from sequence data. P10 sequences do not show any distinct properties that indicate whether or not they form dimeric, trimeric or tetrameric coiled-coils. Coiled-coils can form higher order structures known as α-helix sheets (where coiled-coil interactions occur between several helices arranged side by side) and α-helix cylinders (α-sheets rolled up to form a tube). These structures are identi-fiable by the presence of multiple, overlapping, offset heptad repeats (62). P10 does not appear to possess these sequence patterns.
It would be interesting to model the structure of the coiled-coil domain in silico using the BEAMMO-TIFCC software described by Offer et al. (45) to visualise the effect of the stammer and the presence of the coil breaking residues proline and glycine in the hinge region on the organisation and orientation of the coiled-coil. It may even be possible to determine the higher order organisation of the coiled-coil interaction, such as; does it form a dimeric, trimeric or tetrameric oligomer, is the interaction anti-parallel or parallel, and do P10 oligomers align in frame or staggered? Modelling the potential interactions between two P10 helices in silico, or exploring the possibility of uti-lising protein structure determination techniques, might serve to answer some of these questions.
-
TEM studies have implicated the carboxy-terminal regions (particularly the basic region) as being in-volved in the aggregation of P10 to form FBs (56) and immuno-fluorescence studies suggest deletion of the basic and proline rich regions may alter the shape of P10 aggregates (18). The basic region has been referred to as the potential MT interacting region (46, 58) because MT interacting domains of many microtubule associated proteins (MAP) are positively charged (34, 36, 43, 44). In particular, P10 has been compared to Tau, the major MAP of neuronal cells responsible for stabilising MTs in neurite outgrowths (21). This com-parison originated from a study by Cheley et al. (9), which found that when phosphorylated P10 showed similar microtubule bundling properties to Tau over-expressed in insect cells.
Several MAPs (Tau, MAP2 and MAP4) share a number of structural features. They consist of an acidic amino-terminal projection domain and a carboxy-terminal basic region (30). The basic region contains the microtubule-binding property and is thought to interact with the mainly negatively charged carboxy-terminus of the tubulin microtubule subunit. A proline rich region plays an important role in this interaction. This interaction has been extensively characterised for Tau and has been described by a "Jaws" model where the weak microtubule association of a series of tandem repeats is stabilised by strong microtubule interaction of domains at either ends of the repeats (47). One of these strongly interacting domains is the proline rich region. Because of the similarity of the properties of MAPs with C-terminal domain of P10 it has often been thought that P10 may interact with MTs in a similar fashion. Within the MAPs mentioned above the proline rich region shows a strong positive charge. This differs significantly to P10 where there appears to be a strong negative charge associated with the proline rich region and the positive charge is located outside of the proline rich region. The basic region does not seem to always be required as several strains of BmNPV have a truncated P10 that lacks this region, and both Maruca vitrata nucleopolyhedrovirus (Mavi-NPV) P10 and HaEPV FALPE lack an extensive basic region. While P10 shares a number of features with Tau, including a coiled-coil domain, the difference in size, organisation and chemical properties of the potential microtubule interacting domains is too great to conclude that the carboxy terminus of P10 is analogous to the MT interacting domain of Tau or that P10 interacts with microtubules in a similar way to Tau.
The basic domain shows similarities to a KRKK nuclear localisation motif identified in AcMNPV Polh (28). However nuclear localisation may not be the role of this sequence as P10 is found in both the nucleus and cytoplasm, and P10 is small enough to migrate across the nuclear envelope by diffusion (4). Also the previously described fusion constructs comprising the 51 amino-terminal amino acids of P10 fused to LacZ entered the nucleus (59) suggesting a nuclear locali-sation function for the coiled-coil domain.
No function has yet been ascribed to the proline rich region, although there is a suggestion these regions play a role in the P10 mediated disintegration of the nucleus and release of OBs (56, 58, 63). The P10 proline rich region shows similarities to PEST domains. These are protein domains enriched in proline (P) and glutamate (E) (or sometimes aspartate), and associated with serine (S) and threonine (T) residues, which were originally thought to be involved in inducing rapid degradation of the protein containing them (49). These domains have since been shown to interact with Src homology 3 (SH3) domains (22). SH3 domain-containing proteins are known to be extensively involved in signalling pathways that regulate cytoskeleton kinetics and structure, particularly of the actin network (31). Proline is an amino acid, and its side chain forms a closed ring structure with its peptide backbone nitrogen atom. Due to this its Φ dihedral angle is locked at -60˚, limiting its conformational flexibility. Because of this, and the lack of an amino group to contribute to interpeptide backbone hydrogen bonding, stretches of proline residues often adopt a relaxed left handed helical structure known as a poly-proline type Ⅱ (PPⅡ) helix. The lack of hydrogen bonding capacity also prevents proline from being involved in standard secondary structure formation, such as α-helices and β-sheets. Instead prolines are often found on the exposed surfaces of proteins (27). Regions that adopt PPⅡ conformations are recognised by SH3 and other proline recognition domains (PRDs) (37). The consensus motif for sequences recognised by SH3 domains, PXXP (53), can be found in many P10 sequences, but none possess the preferred sequence where the second residue is also a proline. There does not seem to be a consensus for the proline rich region among P10 sequences. Two motifs seem to occur repeatedly. Many P10 sequences show repeats of a three amino acid motif consisting of a proline residue, an acidic residue (glutamate or aspartate) and a small hydrophobic residue (valine, leucine isoleucine), which we tentatively name a Pah (proline-acidic-hydrophobic) motif. Addi-tionally many P10 sequences possess a two amino acid proline-glutamate (PE) repeat motif, sometimes in conjunction with Pah motives. The PE repeat is not very prevalent in the Alphabaculovirus sequences but forms the entire proline rich region of Christoneura fumiferana granulovirus (CfGV) P10 and is also present in the EPV FALPE protein (2). A more detailed bioinformatics and biochemical study is required to further define and characterise this region.
-
One potential role of P10 that has been mapped to the carboxy-terminal region (57) is its role in mediating nuclear or cellular lysis late in infection to release mature OBs. Cells infected with viruses possessing a p10 mutation were observed to have fewer free floating OBs in their culture media when observed by microscopy (56, 63). Williams et al. (63) described this as a delay in lysis of up to 2 weeks. A virus expressing the carboxy-terminal domain of P10 fused to the adjacent p26 orf (a delayed-early gene of unknown function) was reported as inducing pre-mature cell lysis (7, 23), though it is unknown whether this was due to the early expression of the P10 carboxy-terminal domain or due to interference with p26 function. Van Oers et al. (56) failed to see a difference in cell lysis levels between p10 mutant and wild type viruses utilising a lactate dehydrogenase release assay and concluded that the lack of free floating OBs was due to them being retained within intact nuclei. It is possible that the intact nuclei observed by van Oers et al. (56) are identical to the "naked" OB bundles, more correctly described as mature released OB bundles, previously described by us (6) and have in fact been released from their cell and nuclei but remain stuck together. We have observed that these OB bundles are more prevalent when microtubules are disrupted by drugs (6) or when the p10 orf is deleted (Carpentier et al., unpublished results). We suggest that P10 may act as a non-stick surface coating, helping the dissociation of OB bundles into free floating individual OBs, enhancing their dissemination in the environment and aiding in horizontal spread. Further work is needed to confirm this hypothesis.
The very-late time of expression and its intimate interaction with OBs strongly suggest P10 is involved in the occlusion process and the horizontal spread of virus. Bio-assays of viruses with disrupted functions have yielded variable results regarding in vivo patho-genesis and spread. Vlak et al. found that viruses expressing a P10/LacZ fusion protein were almost twice as virulent as wild type (59). On the other hand there are several studies that indicate reduced tran-smissibility and pathogenicity (higher LD50 and LT50 values compared to wild-type) of viruses possessing a P10 disruption or deletion (58) (Patmanidi AL & King F personal communication). These studies have not yet been convincing enough to provide a definite answer. It is not yet known how P10 associates with OBs. This association appears to occur upon release of OBs from infected cells (6) and can be induced to occur earlier through interference of the carboxy terminus by attachment of an epitope tag (Carpentier et al. unpublished results). It is possible that P10 is only weakly associated with OBs and much of it is removed during the OB purification steps preceding bioassay experiments. The OB bundle disaggregation property of P10 needs to be confirmed and disse-mination experiments need to be designed to measure its influence on baculovirus spread.
P10 is a tiny protein yet it seems to be involved in a multitude of functions and displays varying properties over time. How P10 encodes all these functions is not yet understood. Phosphorylation of a carboxy-terminal serine residue is known to enhance the microtubule association of AcMNPV P10 and induce cellular projection formation (9). A baculovirus phosphatase known to have tyrosine and serine/threonine dephosp-horylation properties has been documented associating with specific regions of FBs (38). The role of post translational modifications in controlling P10 functions remains unknown, however in many SDS-PAGE analyses P10 can be seen to migrate as a doublet band indicating different levels of modification. Lee et al. (35) observed a shift in the relative levels of different P10 species over time. A faster migrating species of P10 was detected from 96 hpi onwards and formed the main species of P10 by 120 hpi. Almost all P10 homologues possess potential phosphorylation recog-nition sites for several kinases, and the distribution of these sites is highly conserved (DCJ Carpentier, unpublished results). It is likely that the virus utilises post translational modification to control P10 mediated processes and trigger them at certain times in the cell cycle or the host caterpillar life cycle (in response to certain hormones produced by the caterpillar), or limits certain functions to specific cells. Phosphorylation of coiled-coil proteins is known to induce unfolding of the α-helix and induce protein aggregation (42, 66) contributing to diseases caused by protein aggregate deposits (such as Alzheimer's disease, caused by the abnormal aggregation of Tau in neural tissue). The P10 FB structures, particularly those in the cytoplasm, often appear very irregular and could potentially be deposits of incorrectly folded or associated protein aggregating in the perinuclear region, where they may form a reservoir of P10, which, upon cell lysis, is available for coating released OBs.
-
The role of the baculovirus P10 protein in the baculovirus replication cycle and in the horizontal transmission of infection between host organisms remains a mystery. Several tentative suggestions have been postulated but none have been concretely proven. P10 is tiny in size but is produced in large quantities and appears to dominate the infected cell during the later stages of baculovirus infection. P10 homologues are conserved within the Alphabaculoviruses and a number of other lepidopteran baculoviruses and poxviruses, suggesting it plays an important role in the life cycle of Lepidopteran viruses. Contradictory to this, the current accepted view is that P10 is dispen-sable for baculovirus infection, both in vitro in cell culture and in vivo in caterpillar hosts. It seems extremely wasteful for baculoviruses to invest such large amounts of resources into i) producing large quantities of the protein during the replication cycle, and ii) evolutionarily maintaining the gene. It is highly likely that P10 plays an unknown, extremely important but non critical role. Viruses that possess this protein must have a strong evolutionary advantage over viruses that do not, otherwise the gene would not be evolutionarily conserved. Baculoviruses that possess a P10 homologue display significant differences in the characteristics of infection progression, both at the level of tissue tropism (showing a wider spread of infection within infected larvae) and of cytopathology (the Betabaculoviruses, which generally lack a P10 homologue, induce nuclear envelope disintegration early in infection and replicate in a mixed nuclear cytoplasmic area of the cell) (16). In recent years interest in P10 has resurfaced (6, 17, 18, 46). We are very close to unravelling the mystery of P10.













 DownLoad:
DownLoad: