-
Transmissible gastroenteritis virus (TGEV) causes acute infection in swine of all ages, and the disease is characterized by severe enteritis and diarrhea. These symptoms are especially severe in newborn piglets less than 2 weeks old, frequently leading to death with mortality rates up to 100% [9, 15, 17]. TGEV was first reported in the United States in 1946 and has now been reported in many other countries, including China [10, 23, 24]. TGEV is now prevalent throughout China and is a cause of serious economic losses.
TGEV is a member of the Coronavirus genus and the Coronaviridae family. Its genome consists of a single molecule of positive-sense, single stranded RNA, 28.5 kb in size, which is transcribed into several subgenomic mRNAs for the production of four structural proteins [Spike protein (S), membrane protein (M), small membrane protein (E) and nucleocapsid protein (N)] and four non-structural proteins (Replicase, 3a, 3b, and Protein 7)[16]. The genes are arranged in the order 5'-rep-s-3a-3b-e-m-n-7-3'[21]. Transcription regulatory sequences (TRSs) including a highly conserved core sequence (CS), 5'-CUAAAC-3', or a related sequence, are present at sites immediately upstream of the TGEV genes[1]. The S glycoprotein makes up the large surface projections of the virion and plays an important role in the attachment of viral particles to host cell receptors, with subsequent penetration into the cell by membrane fusion. The S glycoprotein also stimulates the production of neutralizing antibodies in the host[18]. The M and E proteins are essential for viral envelope formation and release; the M protein can also stimulate the production of interferon (IFN)[12]. The N protein participates in transcription of the viral genome, the formation of the viral core, and packaging of viral RNA[20]. The Coronavirus replicase is a multifunctional polyprotein with helicase and protease activities necessary for the transcription of viral RNA[4, 23]. Four major antigenic sites, A, B, C and D, were characterized on the N terminus of the S protein. Using monoclonal antibodies, the adjacent sites A and B were mapped to a region of approximately 200 amino acids (aa) beginning from residue 506 at its N terminal boundary. The two antigenic sites also overlapped with the S protein domain encompassing aa 522–744 that mediates aminopeptidase N (APN) receptor binding [5, 6, 8]. After continuous passage in cell culture, TGEV isolates gradually lose their virulence and may shift from enteric to respiratory tropism[14]. Attenuation of virulence and tropism shift can also occur in nature, an example being the naturally occurring s and orf3 gene deletion mutant, the Porcine respiratory coronavirus (PRCV), which has both reduced pathogenicity and a predominantly respiratory tropism[14]. Although it is generally accepted that deletions in the spike and orf3 genes contribute to tropism change and attenuation in PRCV, other genes may also be involved. For instance, amino acid mutations in the M protein affect the ability of attenuated Purdue TGEV P115 to induce IFN-alpha, implying a potential role for the M protein in altered host response and virulence[12].
In this study, the complete genome of TGEV TS strain isolated in Gansu province has been cloned. In addition, the virus was continuously propagated in different cell lines and the 3' end genome sequences were also cloned. These data are useful for further study of the molecular structure of the TGEV strains prevalent in China, and the characterization of the long-term TGEV genome stability in different host cells.
HTML
-
TGEV strain TS was isolated from a suburb in Gansu province, China. Swine testicle (ST), PK-15 and IBRS cells were grown as monolayers in DMEM (GIBCO, USA) containing 10% fetal calf serum (GIBCO, USA) and 5% CO2 in air. Samples were obtained by passaging the first TGEV field isolate 10 times in swine intestine and named TS-ST.
-
Total RNA was isolated from infected cell samples using a RNA extraction kit (Qiagen, Germany) following the manufacturer's instructions.
-
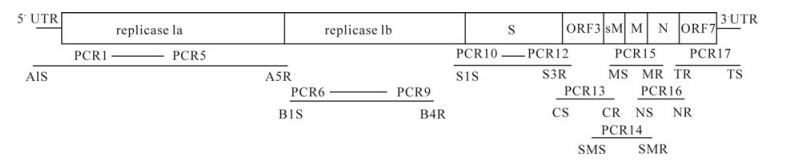
The RT-PCR amplifications were carried out using seventeen primer sets (Table 1) and an RT-PCR amplification Kit (Toyobo, Japan) according to the manufacturer's instructions (Fig. 1).

Table 1. Primers used in cloning the full-length TGEV TS genome
-
RT-PCR products were purified using a Gel Extraction Mini Kit (TaKaRa) and were ligated to the pMD18-T plasmid (TaKaRa). Competent E.coli JM109 were transformed with the recombinant vector. Positive clones were sequenced by the TaKaRa Biotechnology (Dalian) Co. Ltd.
-
Phylogenetic analysis was performed for the TGEV TS strain compared to other coronavirus strains retrieved from GenBank. The sequencing data were aligned using the Clustal W method (Lasergene program version 7.1, DNASTAR Inc. Madison, WI). Phylogenetic trees were constructed for the s and replicase genes using the bootstrap N-J method in the DNASTAR program using 1000 bootstraps. The following sequences, taken from GenBank, were included in the analysis: Feline infectious peritonitis virus (FIPV, AY994055), Porcine epidemic diarrhea virus (PEDV, AAK38661), Human coronavirus 299E (HCoV 229E, Q05002), Severe acute respiratory syndrome virus (SARSV, AAP41036), Avian infectious bronchitis virus (AIBV, P27920), Murine coronavirus (MCoV, P16342), Bovine coronavirus (BCoV, Q91A29), Porcine respiratory coronavirus (PRCV ISU-1, DQ811787), TGEV/Miller M6 (DQ811786), TGEV/Miller M60 (DQ811786), TGEV/Virulent Purdue (DQ811789), TGEV/Pur46-MAD (NC002326), TGEV/SC-Y (DQ443743), TGEV/Purdue P115 (DQ81178/US), TGEV/attenuated H/ EU074218/ China, TGEV/Purdue P-15 (X06371), TGEV/HN-2002 (AY587882), TGEV/FS772-70 (X53128), TGEV/TSX (DQ001167), TGEV/96-1933 (AF104420), TGEV/TO14 (AF302263), TGEV/TH-98 (AF494337).
-
TS was plaque-purified and passaged in ST, PK-15 and IBRS cells 50 times. The clones were named TS-ST50, TS-PK50 and TS-IB50 respectively. The 3' end sequences of the TS-ST50, TS-PK50 and TS-IB50 genomes were amplified and cloned.
Viruses and Cells
RNA Extraction
RT-PCR
Cloning and sequencing
Phylogenetic analysis
Characterization of TS after continuous passage in cultured cells
-
The complete genome of the TGEV TS strain comprised 28 541 nucleotides (nt), excluding the poly (A) tail. The 5' end two-thirds of the genome nt 1-20368 contained a 314 nt non-translated region (NTR) and an open reading frame (ORF) encoding the viral RNA-dependent RNA replicase. The remaining 8 173 nt incorporated the structural genes orf3 and orf7, and a 3'NTR. A small ORF (uorf) with the potential to code for a 3-amino acid peptide at the 5' NTR of the genome, from nucleotide position 99-108 (5'-ACUUUUAAAG-3'), was also found. A conserved sequence was present at different distances from the initiation codon of each gene, and the 3' end of the TGEV genome had a poly (A) tail of undetermined length. The complete TGEV TS genome sequence has been submitted to NCBI and has been given the GenBank Accession Number, DQ201447.
-
The replicase genes were designated genes 1a and 1b, and share 43 overlapping nucleotides. The replicase 1a gene was predicted to encode a protein of 4017 aa while the replicase 1b gene was predicted to encode a protein of 2 680 aa. Nucleotide sequence analysis revealed that there were no deletions or insertions in the replicase genes of all TGEV strains. Compared with the TGEV sequences, PRCV ISU-1 encoded a replicase 1b protein of identical length and a replicase 1a with a 3 amino acid deletion (Table 2).

Table 2. Length in amino acids of predicted structural and nonstructural proteins of the seven TGEV isolates and PRCV-ISU-1
TGEV TS strain orf3a and orf3b genes were predicted to encode proteins of 72 aa and 244 aa, respectively, identical to the corresponding Miller M60 genes. Compared with other TGEV strains, a 3 nt insertion present in the TS, as Miller M6 and Miller M60 strains led to one amino acid (histidine) being inserted at the end of orf3a. No deletions or insertions were found in the TS orf3b gene compared to other strains except for Miller M60. A 531 nt deletion in the orf3b gene of Miller M60 resulted in a truncated 3b protein of 67 aa in length. Meanwhile, a 184 nt deletion in the orf3a gene of PRCV-ISU-1 disrupted the predicted open-reading frame of its encoded protein, and a 117 nt deletion in the orf3b gene predicted a shorter nonstructural protein 3b compared to other TGEV strains. The TGEV TS strain orf7 gene was predicted to encode a protein of 78 amino acids, and no insertions or deletions were found compared with other TGEV strains and PRCV ISU-1 (Table 2).
-
The nucleotide sequence of the TGEV TS strain s gene was 4 350 nt in length, encoding a predicted protein of 1 449 amino acids. As such, it was identical in length to the equivalent proteins of the Miller M6 and virulent Purdue strains. Compared with the TGEV M6 strain, a 3 nt deletion was found in the s gene of Miller M60 at position 2 385 -2 387, which predicted a spike protein of 1 448 amino acids in length. A 6 nt deletion in the s genes of Pur46-MAD and Purdue P115 predicted that their spike proteins were 2 amino acids shorter than that of the TGEV TS strain. Sequence analysis confirmed the previous report of a 681 nt deletion in the 5'end of the s gene of PRCV-ISU-1[14].
There were no deletions or insertions in the e and n genes of all the TGEV strains. The deduced amino acid sequences of the e and n genes of TGEV and PRCV ISU-1 were 82 and 382 amino acids in length, respectively. The m gene of M60 had a 6 nt insertion compared with other TGEV strains, predicting a membrane protein 2 amino acids longer than in the other strains. PRCV-ISU-1 had a 3 nt deletion in the m gene compared with TGEV TS strain, predicting an M protein of 261 amino acid in length.
-
The nucleotide and predicted amino acid sequences of each region (replicase, S, 3a, 3b, E, M, N and ORF7) of TGEV TS, other TGEV reference strains and PRCV ISU-1 were compared. Protein N exhibited the highest amino acid similarity (97.9% -99.7%), protein E showed the lowest amino acid similarity (96.4% -100%), while the amino acid homologies of proteins S and M were 97.8% -99.6% and 97.3% -99.2%, respectively. In the non-structural protein regions, the replicase was highly conserved (97.6% -99.7% amino acid similarity), while the 3b (95.7% -98.8% amino acid similarity) and ORF7 (93.8% -100% amino acid similarity) proteins were less conserved. The highly variable protein 3a showed the lowest conservation (87.8 % -97.3% amino acid similarity). TGEV TS ORF7 shared 100% homology, both in nucleotide and amino acid sequences, with the attenuated strain H also isolated in China (Table 3).

Table 3. The nucleotides and amino acids homology of genomes compare the TGEV TS to other TGEV
-
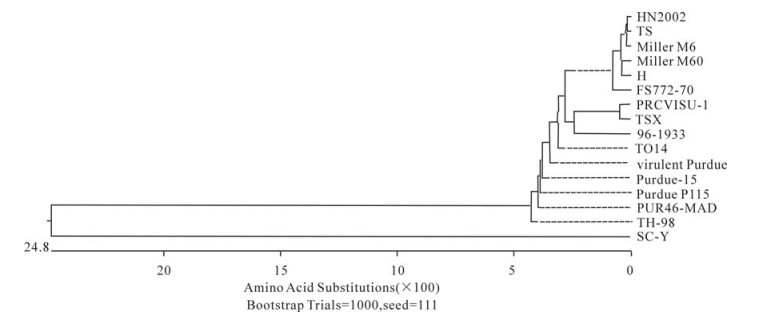
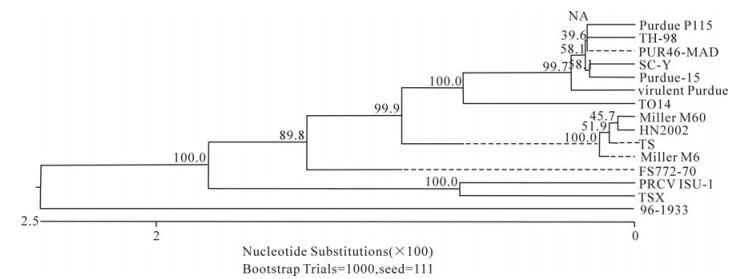
In order to determine the relationship between the TGEV TS strain and other Coronaviruses, a phylogenetic tree based on the Coronavirus replicase amino acid sequences was constructed. The data indicated that all TGEV strains and PRCV ISU-I clustered into one group, and that the TGEV strains were most closely related to FIPV. SARS, which was discovered in China in 2003, had a closer relationship with MHV and BCoV, which belong to the Coronavirus group 2 (Fig. 2). The tree based on the deduced amino acid sequences of the TGEV replicase genes revealed that the strains compared were divided into 2 groups: the Miller group and the Purdue group (Fig. 3). The tree based on the nucleotide sequences of the TGEV s genes showed that all the TGEV strains analyzed were divided into 3 clusters, and that the TGEV TS strain shared the closest relationship with Miller M6 and HN2002 (Fig. 4). Another phylogenetic tree based on the nucleotide sequences of the s genes of TGEV strains and PRCV ISU-1 was also constructed. This revealed that all TGEV strains analysed, except LZC, were divided into 3 clusters (Fig. 5). All phylogenetic trees demonstrated that the TGEV TS strain shared the closest relationship with Miller M6, while PRCV was most closely related to TSX of the British group of coronaviruses.
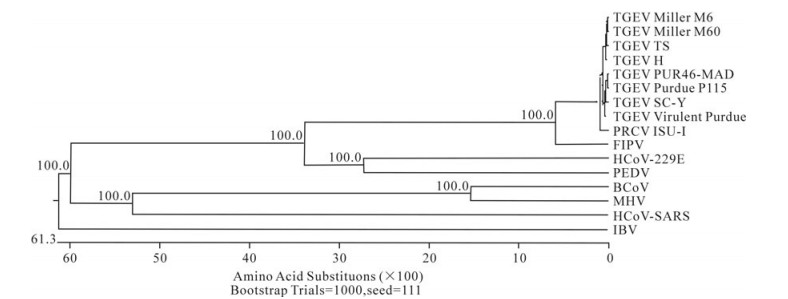
Figure 2. Phylogenetic tree with bootstrap N-J method constructed based on the TGEV strains and other Coronavirus replicase amino acid sequences.
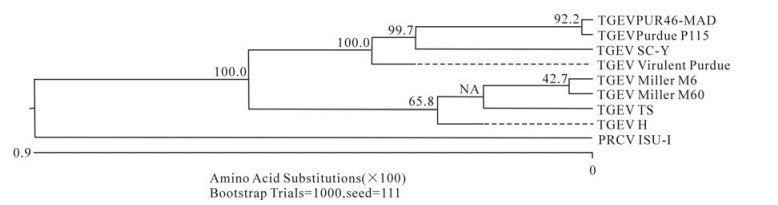
Figure 3. Phylogenetic tree with bootstrap N-J method was constructed based on the TGEV TS and other TGEV strains replicase amino acid sequences.
-
After passaging in swine testicle (ST) cells for 10 generations, cytopathic effects (CPE) were observed after 48 h in culture, and the genes could be cloned from the cells that displayed CPE. CPE was also observed 48 -72 h from the start of continued passage in PK-15 and IBRS cells for 10 generations, but the genes could not be cloned from the cells that displayed CPE. After passage for 50 generations, the 3' ends of the viral genes could be cloned from ST and PK-15 CPE cells, but the genes could not be amplified from IBRS cells that displayed CPE.
The RT-PCR results from ST (TS-ST50) and PK-15 (TS-PK50) cells cultured with TS for 50 passages indicated that there were no nucleotide deletions or insertions at the 3' end of the TGEV genome. However, there were nucleotide mutations in the s and orf3 genes, with TS-ST50 showing less variation than TS-PK50 (Table 4).

Table 4. The s and orf3 nucloetides mutations from TS to TS-ST50 and TS-PK50
Complete genome sequence of the TGEV TS strain
The non-structural genes
The structural genes
Homology comparisons
Phylogenetic tree analysis
Characterization of TS genome stability in long-term cell culture
-
Eight complete nucleotide sequences of the TGEV genome have been reported in GenBank, of which three were collected in China. Analysis of the full genomic sequence of the TGEV TS strain can contribute to the understanding of the genetic structure, diversity, evolution and, in particular, the prevalence of these coronaviruses in China.
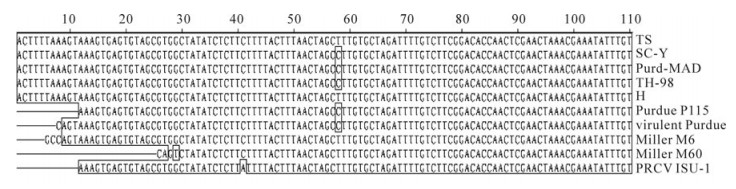
The uORF is present in all Coronaviruses and plays an important role in virus replication and packaging. The uORF sequences of HCoV-299E and IBV were found to be eleven codons in length, while those of MHV and PEDV were eight and twelve codons, respectively. Meanwhile, the uORF of TGEV encodes a peptide of only three-amino acids[11]. In this study, we demonstrated that the TS strain has the same three-amino acid uORF peptide as other TGEV strains, indicating that uORF is highly conserved in TGEV. Moreover, most of the 5' NTR sequences containing uORF were conserved, except for the first few nucleotides. In this study, we used two sense PCR primers beginning with the sequences "ACTTTTAAAGTAAAGTGAGT" and "GCCAGTAAAGTGAGTGTAGC", which corresponded to the Pur46-MAD and Miller M6 sequences, respectively. The PCR results showed that only the former primer could amplify the 5' NTR of the TGEV TS strain, indicating that the TGEV TS strain M6 sequence should start with the "ACTTTTAAAGTAAAGTGAGT" sequence found in the Pur46-MAD strain. This was confirmed by 5' RACE (data not shown). Alignment of all TGEV sequences showed that they were highly conserved except for nucleotide deletions in some strains (Fig. 6). We speculate that incomplete sequencing is responsible for the presence of these deletions, and propose that the first several nucleotides of the 5' NTR are conserved among the TGEV strains. The length of the poly(A) tail of the TGEV genome was not confirmed because it has not been possible to amplify it accurately by PCR, although a poly(A) sequence of 9 residues has been cloned by 3' RACE (data not shown). Whether this sequence corresponds to the true poly(A) tail remains unknown. Nevertheless, minigenomes and full length infectious RNAs with a poly(A) tail of 24 residues have been constructed and replicate efficiently, indicating that 24 A residues are sufficient for TGEV RNA replication [2].
The principal component of the Coronavirus genome is the replicase gene, which contains two large open reading frames, orf1a and orf1b, from which two products are translated. The latter is synthesized via a ribosomal frameshifting mechanism, facilitated by a pseudoknot structure[7]. orf1b sequences in two Chinese TGEV strains, TS and SC-Y, were predicted to encode a protein of 2 678 aa. In fact, because the ribosomal frameshifting mechanism was not accounted for, two amino acids were deleted in ORF1b of TS and SC-Y. In common with other TGEV strains, the predicted orf1a sequences of TS and SC-Y extended from nucleotide 315 to 12054. This resulted in a 4017-codon orf that overlapped orf1b from nucleotide 12 011 to nucleotide 20 368, with the capacity to encode a polypeptide of 2680 amino acids (Fig. 7).

Figure 7. Nucleotide and amino acid sequence of the ribosomal frameshifting region. The first two line is the nucleotides and a.a. of ORF1a, the second two line is the nucleotides and predicted a.a. of ORF!ab that had been in the GenBank, the third two line is the nucleotide and predicted aa of ORF1ab of ribosomal frameshifting, the nucleotides with underline is the ribosomal binding site, the nucleotides with box is the insertion of ribosomal frameshifting.
The orf3 and s genes have been hypothesized to be important in the virulence and pathogenesis of TGEV infections[19]. Analysis of TGEV ORF3 sequences revealed that TS had equal numbers of ORF3 nucleotides and amino acids as the virulent Miller M6 strain. The G residue at nucleotide position 655 of the spike protein was essential to maintain enteric tropism of the TGEV strain PUR46-MAD, and mutation at this nucleotide caused a shift from enteric to respiratory tropism[3]. The T to G mutation at nucleotide 1 753 of the s gene is regarded to be a signal of attenuation[25]. Analysis of the s gene sequence of the TGEV TS strain revealed a G at nucleotide 655 and a T at nucleotide 1 753 indicating that TS was a virulent strain, also demonstrated by animal experiments (data not shown). A 6 nt (nt 1 122 to 1 127, aa 375 -376) deletion is present in the attenuated TGEV Purdue P115 and Pur46-MAD spike genes compared to the virulent Purdue strain, and so this deletion was also thought to play a role in attenuation[18]. Comparison of the attenuated Miller M60 strain with the virulent Miller M6 strain showed that no deletion was present at nucleotides 1 122 to 1 127, but there was a deletion at nucleotides 2 386 -2 388 in the s gene. This suggested that the deletion in the s gene may affect virus virulence, but this is uncertain from the position of this particular deletion. The TGEV TS S protein possessed the same number of amino acids as the Miller M6 and virulent Purdue strains, suggesting that TS is also a virulent strain. Amino acid mutations in the M protein affect its ability to induce IFN-alpha, and so the M protein has a potential role in TGEV virulence. The m gene has been regarded as a highly conserved TGEV gene, but comparison of the lengths of TGEV m genes revealed that the Miller M60 gene (789 nucleotides) encoded a 263 aa polypeptide, the TGEV gene encoded a 262 aa polypeptide, and the PRCV ISU-1 gene encoded a 261 aa polypeptide. The aa insertion in TGEV is suggestive of the M protein being subject to environmental selection pressure because it affects a coat protein. Although the ORF7 nucleotide deletion of TGEV had been considered to affect virulence[13], we found no deletion among the TGEV strains and PRCV ISU-1.
Homology and phylogenetic tree analysis showed that TGEV TS had the closest sequence relationship to the Miller M6 strain. All the compared TGEV strains were divided into 3 clusters in the phylogenetic tree based on the s gene nucleotide and amino acid sequences. These were the American Miller cluster, American Purdue cluster and British cluster. The attenuated strain H from China had the closest relationship with Miller M60, while another Chinese strain, SC-Y, belonging to the Purdue cluster was separated into an independent group based on the S amino acid phylogenetic tree. Analysis of SC-Y gene revealed that one nucleotide mutation generating a termination codon appeared at aa residue 924. This may have been due to a sequencing mistake because no such mutations have been described in coronavirus s genes. Surprisingly, the orf3a genes of viruses in the Miller cluster were 72 aa in length, while in the Purdue cluster they were 71 aa in length. Therefore, the nucleotide number of the orf3a genes may differentiate the Miller cluster and Purdue cluster strains. Homology analysis indicated that PRCV ISU-I had the closest relationship with the Miller strains at both the nucleotide and protein levels. The orf3 gene of the Miller strains also had a high frequency of mutations and deletions, suggesting that a Miller-like strain was the ancestor of PRCV-ISU-1[25]. In this study, we confirmed that PRCV-ISU-1 had a closer relationship with Miller strains than the Purdue strains. The TS strain shared 100% aa homology with PRCV-ISU-1 in the e gene, indicating that TS may have the closest relationship with the PRCV-ISU-1. Interestingly, the phylogenetic tree based on the s gene sequence showed that PRCV-ISU-1 had the closest relationship with TSX, which belongs to the British cluster.
As described above, we have determined the entire genomic sequence of the TGEV TS strain isolated from the Gansu province of China. We characterized the TS strain by following its passage in three different cell types typically used for the culture of TGEV. As expected, ST cells were most sensitive to the presence of the TGEV virus, as indicated by the presence of cytopathic effects (CPE) after 48 h in culture. Although CPE were obvious in IB cells passaged for 50 generations, TGEV genes could not be cloned by RT-PCR, suggesting that maybe the virus can not be propagated in the cells. This indicated that IB cells were not suitable for the culture of the TGEV virus. The 3'end genome sequences of TS-ST50 and TS-PK50 showed no nucleotide mutations in the m, n, e and orf7 genes. However, the s and orf3 genes of TS–PK50 had more nucleotides mutations than in TS-ST50. Surprisingly, the T residue at position 1753 of the TS s gene mutated to G in TS–PK50. This is regarded as a signal of attenuation, indicating that TGEV TS virulence could be attenuated by continued passage in PK-15 cells.













 DownLoad:
DownLoad: