-
Hepatitis B virus (HBV) possesses a partially double-stranded DNA genome that encodes four overlapping open reading frames (ORF), including C, X, P, and S [12]. The S-ORF encodes three surface proteins, L, M, and S-protein (HBsAg), through alternative translation initiation (ATI) from three in-framed start codons [6, 15, 17, 18].
Naturally occurring mutations of surface proteins identified by clinical screening have been extensively documented in recent years [1, 3, 7, 14, 19]. However, certain amino acid residues bearing critical function for virus viability are highly conserved and mutations rarely occur among the various HBV genotypes, which makes clinical screening insufficient to address the importance of these highly conserved residues. In this report, site-directed mutagenesis of highly conserved residues in the S-domain revealed two methionines, mutations of which decreased HBV genome replication level without compromising the overlapping p-gene product.
HTML
-
Hepatoma cell line Huh7 cells maintenance and plasmids transfection were performed as previously described [10].
-
All HBV genome-derived sequences were cloned using polymerase chain reaction (PCR) from pAdr-1 [4], sequences of primers used in this study are listed in Table-1.
To introduce mutations into the S-ORF in pHBV1.3 [9], two-step PCR was performed. The first round of PCR used primer sets 1.3up/R75 and F75/1.3dn, respectively. The subsequent PCR products were mixed in equal molar as template and amplified using primer set 1.3up/1.3dn to generate M75T mutant HBsAg encoding gene, by converting ATG to ACG. The final PCR product was digested with EcoR I and Spe I and replaced the corresponding fragment of pHBV1.3 to generate pHBV1.3-75 (Fig. 1A). Similarly, pHBV1.3-103 was generated with primer sets 1.3up/ R103 and F103/1.3dn (Fig. 1A). pHBV1.3-75-103 bearing M75T and M103T was constructed as pHBV1.3-103, except pHBV1.3-75 was used as initial template (Fig. 1A).

Figure 1. Plasmid constructs. A: pHBV1.3 and derived plasmids encoding M75T or/and M103T within HBsAg. B: pSG and derived plasmids encoding M75T or/and M103T within HBsAg. C: Plasmids expressing truncated HBsAg, in which coding sequences of either 1-74 (pS75) or 1-102 (pS103) AA on N-terminal of HBsAg were removed.
To label HBsAg with EGFP, HBsAg encoding genes were amplified by PCR using primer set Gup/Gdn. The resultant fragment was cloned in frame into pEGFP-N1 (Clontech) between NheI and ApaI, to generate pSG harboring egfp-fused HBsAg (Fig. 1B). To introduce mutations into pSG, site-directed mutagenesis was performed. Plasmids pSG-75, pSG-103, and pSG-75-103, each bearing either M75T, M103T or dual mutations, were generated using similar methods as described above, with primer sets Gup/R75, F75/ Gdn, Gup/R103, and F103/Gdn (Fig. 1B).
Two pcDNA3.1-based (Invitrogen) vectors expressing truncated HBsAg with the HA tag were generated using primer sets 3.1-75/3.1dn and 3.1-103/3.1dn, which spanned HBsAg from M75 (pS75) or M103 (pS103) to the C-terminal, respectively (Fig. 1C).
All the plasmids were verified by sequencing.
-
HBV replicative intermediates were purified from intracellular core particles of cells transfected with indicated plasmids, and submitted to Southern-blot assay as previously described [20].

Table 1. Primers used in this study
-
HBsAg and HBV e antigen (HBeAg) in the supernatant or cell lysates from indicated HBV-bearing plasmids transfected Huh7 cells were analyzed by ELISA kit (Kehua) according to the manufacturer's instructions.
-
Huh7 cells cultured on coverslips were transfected with indicated plasmids. At 48 hour post transfection (hpt), cells were submitted to fluorescent microscopic assay by using an Olympus IX51 inverted microscope.
-
1163 HBsAg sequences (genotypes from A to H) were retrieved from the Hepatitis virus database (http://s2as02.genes.nig.ac.jp/), and aligned to pAdr-1. The conservation of ATG start codons in front of each putative ORF on the S-ORF was calculated using a Perl script (available upon request).
-
Huh7 cells transfected with indicated plasmids were harvested at 48 hpt. 20 μg total cell lysates of each transfected cells were submitted to Western blot with anti-GFP antibody (Sigma).
Cell culture and transfection
Plasmid constructs
Southern-blot
Enzyme-linked immunosorbent assay (ELISA)
Fluorescent microscopy
Sequence analysis
Western-blot
-
The role of surface proteins in HBV envelopment and secretion has been described [2, 13]. In this research, M75T and M103T were introduced to the surface proteins to examine their impact on HBV genome replication and viral antigens secretion.
Compared to pHBV1.3 transfected cells, Southern blot indicated a significant reduction in de novo synthesized relaxed circular (RC) and single-stranded (SS) HBV DNA in cells transfected with either pHBV1.3-75 or pHBV1.3-75-103 (Fig. 2A), as compared to pHBV1.3. M103T alone had a less profound effect. Since all the three mutations introduced to S-ORF generated silent mutations to the overlapping polymerase gene, the impaired HBV replication capability was attributed to the mutations in the S-ORF. Moreover, the HBsAg secretion was significantly suppressed in the mutants bearing the M75T mutation (Fig. 2B). The HBeAg secretion of each mutant was not changed signifi-cantly (Fig. 2C), which indicated the function of the cellular system in respect to secretion of HBV-associated antigens remained unaffected.
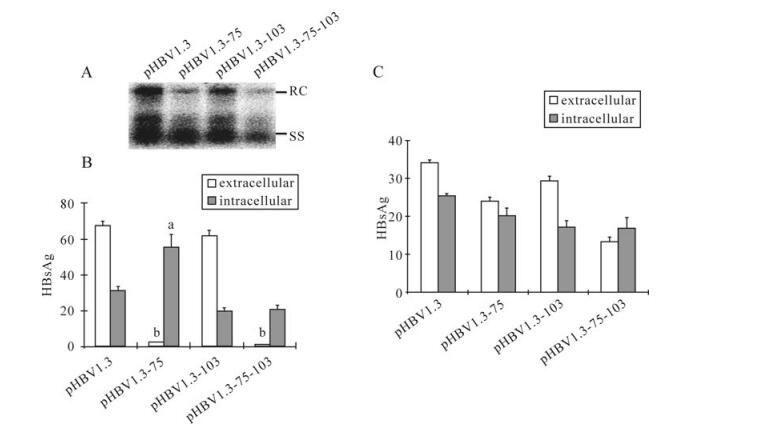
Figure 2. The effect of M75T and M103Ton HBV genome replication and HBsAg secretion. A: Analysis of intracellular encapsidated HBV DNA. Indicated plasmids were transfected to Huh7 cells. At 96 hpt, encapsidated viral DNA was harvested and submitted to Southern-blot hybridized by 32P-labeled HBV DNA probes. B: Analysis of HBsAg. After transfection with indicated plasmids, supernatants and cell lysates were collected at 48 hpt and submitted to ELISA. The letters indicate statistical significance (aP < 0.05, bP < 0.001, Bonferroni-Dunn post-hoc test) between pHBV1.3 and the corresponding mutant. Data are means ± SD. C: Analysis of HBeAg. Done as in B.
-
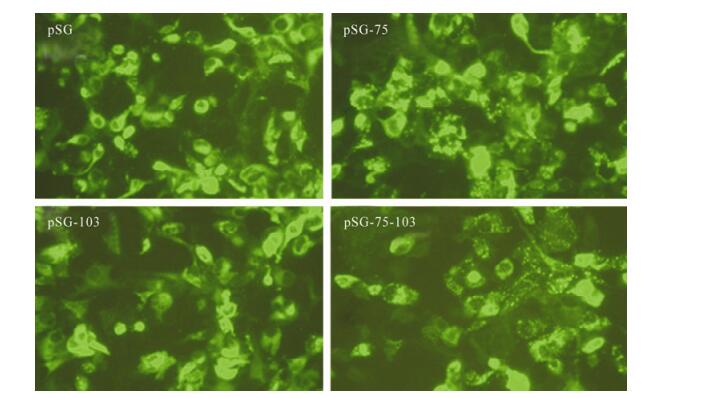
In the cells transfected with pSG-75 or pSG-75-103, EGFP-fused HBsAg exhibited a puncta-like pattern, which was in contrast to the uniform diffusion pattern in pSG or pSG-103 transfected cells (Fig. 3). A previous report observed that a L77R mutation in HBsAg resulted in HBsAg accumulating in the lumen of endoplasmic reticulum (ER) and Golgi complex [2]; the punctated fluorescence pattern found here suggested the M75T mutation might lead to similar HBsAg accumulation in the ER and Golgi lumen and prevent HBsAg achieving extracellular secretion.
-
Although the above experiments demonstrated mutations in surface proteins contributed to impaired HBV replication, whether the surface proteins were directly involved in HBV replication remained ambiguous. One possible scenario is M75 and M103 encoded by ATG could potentially initiate novel truncated surface proteins (TSPs) synthesis by ATI. In this mechanism, the surface proteins may not directly participate in HBV replication regulation, but TSPs initiated from M75 or M103 may be functionally important for HBV replication.
Sequence analysis indicated the ATGs encoding M75 and M103 were highly conserved among various HBV genotypes (Fig. 4A). To test whether M75 and M103 were capable of acting as ATI sites, egfp-fused HBsAg encoding plasmids were transfected into Huh7 cells. Total cell lysates were submitted to Western-blot and probed with anti-GFP antibody. Surprisingly, besides the major bands of HBsAg-EGFP, the blot of pSG exhibited two minor bands of 46kD and 43kD (Fig. 4B), the molecular weight of which were in accordance with the putative proteins translated from M75 and M103 respectively. Meanwhile, the minor bands of 46kD and 43kD disappeared with the M75T and M103T mutation respectively (Fig. 4B), which suggested M75 and M103 may act as ATI sites initiating novel TSPs synthesis when HBsAg is over-expressed. Accordingly, both the 46kD and 43kD bands disappeared with the M75, 103T dual mutation (Fig. 4B), which further confirmed M75 and M103 accounted for the novel TSPs. Similar results were obtained by testing EGFP-fused L and M proteins (data not shown).
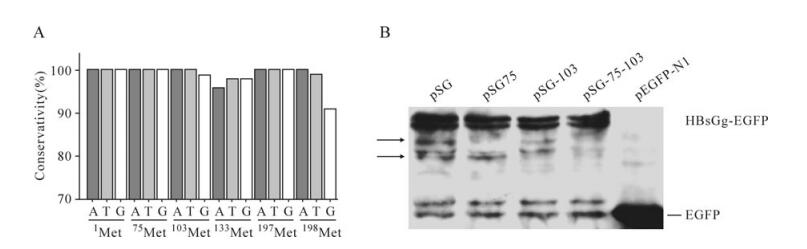
Figure 4. A: Nucleotide conservation of ATGs in HBV S ORF. B: Western-blot assay of HBsAg and related mutants. At 48 hpt, Huh7 cells transfected with indicated plasmids were harvested and submitted to western blot. The arrows indicate the minor bands, for which molecular weight was in accordance with putative alternative translation starting from M75 and M103.
-
To explore the possibility of putative TSPs rather than surface proteins accounting for the impaired HBV replication, a series of trans-complemented rescue attempts were performed by co-expression of TSPs with the corresponding replication-defective HBV mutant.
Southern-blot results indicated HBV replication of pHBV1.3-75, pHBV1.3-103, or pHBV1.3-75-103 remained at a low level even after co-expression of the corresponding TSPs (Fig. 5). This phenotype excluded the possibility that the putative TSPs accounted for the impaired HBV replication induced by M75T and M103T mutations, albeit their virtual existence in the context of HBV genome remained obscure, and indicated that surface proteins were directly involved in regulation of HBV replication.
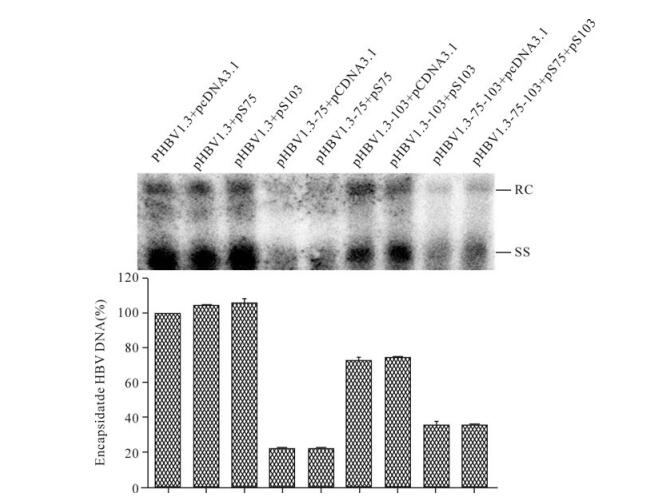
Figure 5. Attempts to rescue HBV replication with TSPs. Huh7 cells were co-transfected with indicated plasmids. Done as in Fig. 2A. Lower panel showed the quantification of Southern-blot data from upper panel. The value of the HBV DNA contents in the corresponding mutant transfected cells was normalized to that of pHBV1.3 transfected cells.
-
After insertion as a monomer into the ER membrane, the oligomerization of HBsAg appears to be the next event before the secretion [16]. Some mutations of HBsAg abolish the secretion, but do not affect the oligomerization [22], therefore the mutant can retain the WT HBsAg in the cell by forming intermolecular aggregation. To study whether the mutants bearing M75T also possessed the ability to retain the WT-HBsAg, pHBV1.3-75was co-transfected with pHBV1.3 at the ratio of 4:1. Interestingly, pHBV1.3-75 did not affect WT-HBsAg secretion (Fig. 6), implying M75T might influence the oligomerization of HBsAg.
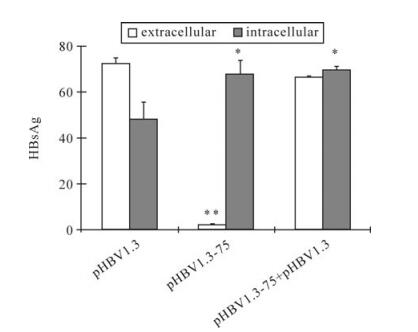
Figure 6. The effect of M75T-carrying mutant on the secretion of WT HBsAg. Key is the same as in Figure 2B. pHBV1.3-75 was cotransfected with pHBV1.3 at the ratio of 4:1.
Mutations of S-ORF decreased HBV genome replication level
M75T altered HBsAg subcellular distribution pattern
TSPs synthesis upon over-expression of full-length surface proteins
TSPs failed to rescue replication-defective HBV mutants
M75T did not affect the secretion of the wild type (WT) HbsAg
-
HBV replication is a complicated process regulated by various host and viral factors [5, 11, 21]. Although the surface proteins mainly exert their functions in assisting virus envelopment and secretion, there are also data indicating they may play roles in the regulation of replication. The knockout of HBsAg by eliminating the initiation codon increased the RC DNA [22]. The A119F of L and the L77R of HBsAg could enhance the maturation of the virus DNA from the SS form to the RC form in the context of the core 97L mutation, possibly by modifying the interaction between the core and the envelope proteins [2]. In this report, we identified two novel mutations in the S-domain that attenuate HBV replication level by decreasing both RC and SS DNA.
M75 and M103 are located in the S-domain and shared by all the three surface proteins. Although we provided evidence that surface proteins were involved in the regulation of HBV genome replication, the respective roles of each surface protein remained obscure. Combined with the results from previous reports that all three surface proteins are dispensable for HBV DNA replication [22], it is possible they play a regulatory role during HBV replication, and the suppression of HBV replication by M75T and M103T is attributable to the orchestral effects of the mutations on L, M and S proteins. Meanwhile, it should also be noted the effects of host factors stimulated by HbsAg arrest cannot be excluded and need further study.
The cytosolic loop (aa29-80) of HBsAg is important for the secretion of HBsAg. The mutations of R73L, L77R, R78A or R79A at this region can lead to the arrest of HBsAg [22]. The mutation (I110M or R169P) which is predicted to be in the ER lumen or the ER membrane can also hamper the secretion of HBsAg [8]. Moreover, M133T of HBsAg is reported to enhance the secretion [22]. In our study, the conserved M75, not M103, was essential for the secretion of HBsAg. Moreover, M75T-carrying mutant did not retain the secretion of WT HBsAg, implying M75T might abolish the secretion by interrupting the oligomerization of HBsAg. Whether the oligomerization of HBsAg plays a regulatory role in HBV replication needs to be further studied.













 DownLoad:
DownLoad: