-
Double-stranded (ds) RNA viruses infect a very wide range of host species, from bacteria to plants and animals. The Reoviridae, one of the most complex families in the dsRNA virus group, consists of at least 15 distinct genera reported to date. Two sub-families, Spinareovirinae and Sedoreovirinae, are currently classified by the International Committee for the Taxonomy of Viruses (ICTV) based on their particle structural organization (Makkay A, et al., 2011). The current structural information of reovirus particles and individual proteins from different genera has revealed an architectural principle common to some seemingly unrelated dsRNA viruses. In addition, it is also recognized that the structural organization of a number of dsRNA viruses placed in different genera also have similar biological functions in virus infection, even though they have widely divergent genome sequences. It is clear that meaningful sequence comparison is possible only between closely related viruses, an d the present methods of bioinformatics analysis are unable to recognize the similarities between different genera based on their sequence alignments only. Studies involving single-particle cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction of reovirus particles have shown that virus particle organization and structures of different virus types correspond to their function in viral infection and replication cycles. Analyses of reconstructed particle images obtained by these techniques have revealed a common evolutionary architectural principle for structurally different reoviruses, such as mammalian orthoreovirus (MRV, a turreted reovirus) or rotavirus (a non-turreted reovirus). In addition, the progress of cryo-EM techniques has fostered the use of structural information to understand the functions of these complex molecular structures. Recent studies have focused on high-resolution 3D structures of reovirus particles produced by cryo-EM, which approach the resolutions seen in structures identified by X-ray crystallography (Zhang X et al., 2008, 2010a; Settembre E C, et al., 2011; Yu X, et al., 2008). Some results determined by near-atomic resolution reconstructions using cryo-EM allow integration of structural a nd functional information into a coherent mechanism for understanding reovirus assembly and entry. Of the resolved 3D structures, the orthoreovirus, aquareovirus, and rotavirus structures have been well studied by cryo-EM. In this review, we summarize current progress on the structural basis of non-enveloped reovirus assembly and entry, mainly focusing on the MRV, aquareovirus, and rotavirus members of the Reoviridae family.
HTML
-
Virus particles in the Reoviridae family appear to have icosahedral symmetry and an overall diameter of approximately 75–85 nm. These viruses comprise a dsRNA genome of 9–12 segments, enclosed by single, double, or triple proteinaceous capsid shells. Generally, mature reovirus particles contain seven or eight structural proteins, depending on the genus. All MRV members have 10 linear dsRNA segments, and the genomic dsR-NAs are grouped into three size classes, commonly referred to as large (L1–L3), medium (M1–M3), and small (S1–S4), based on their electrophoretic mobilities in gels. Hence, their structural proteins are designated in terms of their relative sizes and size classes: λ1, λ2, and λ3; μ1 and μ2; and σ1, σ2, and σ3. Aquareovirus members contain 11 linear dsRNA segments (termed S1–S11), and the virus particles are composed of seven structural proteins, named VP1–VP7 based on the molecular weight of each protein. In rotavirus, the triple-layered protein capsid also encloses a genome of 11 linear dsRNA segments (Seg1–Seg11), and eight structural proteins (VP1–VP8), making up the intact virus particle. Mature reovirus particles contain an inner core and an outer protein shell. The properties of the structural proteins in members of the MRV, aquareovirus, and rotavirus in the Reoviridae family are schematically depicted in Fig. 1.
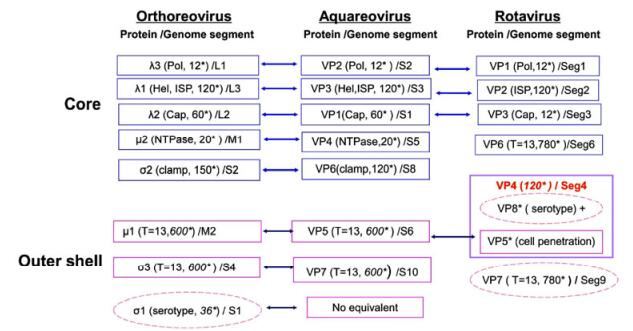
Figure 1. Properties of proteins in mammalian orthoreovirus (MRV), aquareovirus, and rotavirus particles. The reovirus mature particle is composed of a core and an outer shell. The cores of different genera contain inner shell proteins (ISPs) and other proteins responsible for genome transcription and replication, including the RNA-dependent RNA polymerase (pol), capping enzymes (cap), nucleoside triphosphate hydrolase (NTPase), and helicase (Hel). The outer shell proteins play roles during virus infection and cell entry. *Indicates molecular copies of each protein in one particle. Core proteins are shown in blue boxes and outer shell proteins in pink boxes, while the proteins in dashed pink ellipses are serotype-specific antigens.
-
Within the Reoviridae family, the 15 reovirus genera are divided into two sub-families. The cores of the "turreted viruses" group are classified in the Spinareovirinae sub-family, because these virus members have 12 typical icosahedrally pentameric spikes residing on the surface of the core at the five-fold axes. Nine genera belong to the turreted reovirus group; the typical icosahedrally pentameric spikes of aquareovirus is shown in Fig. 2A (top left). Meanwhile, the cores that are distinguished from those of the 'turreted virus' group possess a relatively smooth surface without large surface projections at their five-fold axes, defined as 'smooth reovirus'. Compared with spiked reovirus, it is obvious that the rotavirus core structures have a relatively smooth surface; Fig. 2B(bottom left) shows the 3D reconstruction of a rotavirus core particle. The innermost protein layer of reovirus particles has an internal diameter of approximately 50–60 nm. As schematically shown in Fig. 2A (top right) and Fig. 2B (bottom right), both turreted and non-turreted reovirus core particles contain an RNA replication complex, which includes an RNA-dependent RNA polymerase (RdRp, which functions as a transcriptase and replicase), an NTPase, a helicase, and capping complexes (containing guanylyl transferase and methyl transferase).
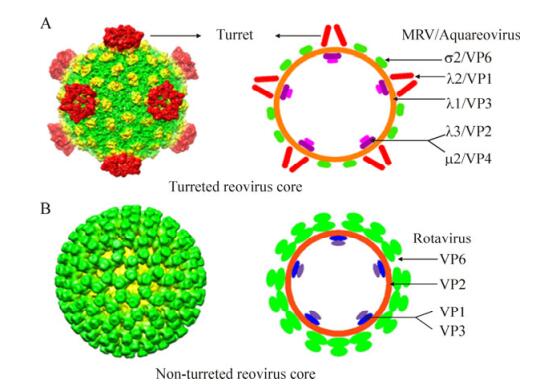
Figure 2. Comparison of two distinct core particle morphologies (spiked and unspiked), present in members of turreted and smooth reovirus in the Reoviridae family. A: Three-dimensional (3D) reconstruction images of turreted reovirus cores (top left panel). On the right is a diagrammatic representation of the MRV and aquareovirus core particles with their core protein locations. B: 3D reconstruction image of the smooth rotavirus core is shown on the bottom left, and on the right is a diagrammatic representation of the rotavirus core particle with each protein location. The 3D images (A, B left) were reproduced from Cheng L P, et al., 2008; Zhang X, et al., 2008. The diagrammatic representations (top and bottom right) were modified from Makkay A, et al., 2011.
The segmented RNA genomes in reovirus are often packaged into a shell formed by an 'inner shell protein' (ISP). The ISPs in both turreted and non-turreted virus groups are comprised of 120 homologous monomers or 60 dimers (Makkay A, et al., 2011). The ISP in MRV is composed of 120 molecular copies of the λ1 protein, which is identical to VP3 in aquareovirus. The typical characteristic of the 'turreted' internal capsid particle (ICP) is a cylindrical pentameric complex, which has been shown to have guanylyl transferase activity, and is also believed to have RNA capping activities, with S-adenosyl-L-methionine binding domains. The capping complexes are ordered structures displayed on the outside of the core shell at the five-fold axes. Five λ2 molecules compose a turret in MRV (Fig. 2A), and in this virus type, the locations of the λ1 (ISP, T=1), λ3 (Pol), and μ2 (NPTase) proteins, which relate to viral repli cation, have been identified as being at the bottom of the five-fold axes (Reinisch K M, et al., 2000; Cheng L P et al., 2008, 2010). Members of other dsRNA genera also possess turret-like structures that are homologous to λ2, such as VP1 in aquarovirus, or turret protein (TP) in cypovirus (CPV). For turreted core structures, three of these genera, Orthoreovirus, Aquareovirus, and Cypovirus, have been well studied by cryo-EM to date (Reinisch K M, et al., 2000; Cheng L P et al., 2008, 2010; Yu X, et al., 2008). The core structures of MRV have been determined at 3.6 Å resolution by X-ray crystallography (Reinisch K M, et al., 2000), while CPV and aquareovirus have been determined at 3.8 Å and 4.5 Å resolution, respectively, by cryo-EM (Yu X, et al., 2008; Cheng L P, et al., 2010). The resolved 3D structures of the cores indicate that the transcriptionally competent ICPs of turreted reovirus members all appear to share a common shape (λ1-like framework), which packages the genome and organizes the polymerization and capping enzymes. In addition to the λ1/VP3 subunits, which compose the core ISP in MRV and aquareovirus, there are σ2/VP6 subunits decorating the frame of the core particles to strengthen each ISP.
The 'non-turret' rotavirusICP is known as a 'double-layered particle' (DLP). Its inner protein layer is comprised of VP2, and the viral protein VP6 builds up the outer protein layers of the DLP. The ISP layer of a rotavirus is assembled from 120 copies of VP2, and trimers of VP6 surround its ISP to form a T=13 (laevo) icosahedral lattice symmetry. VP6 is composed of 780 copies, which are arranged as 260 trimeric morpho-logical units positionedat the local and strict three-fold axes of the icosahedral lattice, which forms the outer surface of the DLP. The assembled genomic dsRNAs, along with the enzymatic complexes and VP2 layer, comprise the rotavirus core. However, the capping enzymes of smooth reovirus (VP3) lie inside the common shell (Fig. 2B), which is in contrast to turreted reovirus. The location of the smooth reovirus capping protein may be in close association with the polymerases. The DLP structures of rotavirus have been determined both by cryo-EM at 3.8 Å nominal resolution (Zhang X, et al., 2008) and by X-ray crystallography at about the same reso lution (McClain B, et al., 2010). The smooth ICP contains an RNA replication complex, such as VP1 (RNA-dependent RNA polymerase), or the capping enzyme VP3 (guanylyl transferase and methylases). These results also suggest that in some cases, the capping enzyme complex possesses an additional set of stabilizing ISP subunits (McClain B, et al., 2010).
Beyond the conserved ISP, 150 copies of σ2 have been found in both MRV and avian orthoreovirus (Reinisch K M, et al., 2000; Zhang X, at al., 2005); however, only 120 copies of VP6 have been found in aquareovirus (Cheng L P, et al., 2008). Analyses of 3D reconstruction by cryo-EM revealed that two conformations of aquareovirus VP6 (termed VP6A and VP6B) located at two different positions in a T=1 asymmetric core. VP6A surrounds each five-fold axis, and VP6B surrounds each three-fold axis. The globular monomer σ2 in MRV binds at three distinct locations within each icosahedral asymmetric unit. There are 60 copies of σ2 at the five-fold axis and another 60 copies at the three-fold axis, which correspond to the localization of the aquareovirus VP6A and VP6B subunits. In addition, σ2 in MRV has been found to have another 30 nodules at the two-fold vertices, which are unoccupied in the aquareovirus core and in CPV (Cheng L P et al., 2008, Yu X, et al., 2008). This fact suggests that the MRV core shell is more stable than that of aquareovirus. In contrast to turreted reovirus, the outer-layer VP6 of rotavirus forms stable trimers with 260 subunits on the outer layer (McClain B, et al., 2010). It is obvious that rotavirus has a different ICP from spiked reovirus. The existence of these two kinds of core surface organization reflects the divergence of structurally related evolution within the Reoviridae family. Previous studies have shown that the inner layers of rotavirus and BTV do not have any decorated element on their core shell. In addition, viruses lacking clamp protein, such as VP2 in rotavirus and VP3 in orbivirus, can self-assemble into icosahedral particles (Labbé M, et al., 1991; Hewat E A, et al., 1992). However, for viruses belonging to the Spinareovirinae sub-family, such as MRV, when core frame protein λ1 is singly expressed in insect cells, no icosahedral particles can form unless clamp protein σ2 is also co-expressed (Kim J, et al., 2002). This suggests that protein σ2 of MRV or its analog in the Spinareovirinae sub-family is indispensable for core shell assembly. Recent 3D reconstruction studies have revealed a high level of structural similarity between the ISPs of viruses in the Spinareovirinae and Sedoreovirinae sub-families, even though there is great divergence at the genome sequence level. The conserved structures largely enhance those functional and enzymatic domains that are responsible for maintaining inner core shell stability and endogenous transcription activity. However, in contrast to their conserved inner capsid shell proteins, the intermediate (e.g., clamping protein VP6/σ2) and oute r protein components of these dsRNA viruses have diverged significantly. The conserved ISP structures remaining in both spiked and smooth reoviruses reflect similar mechanisms for RNA transcription and replication.
-
Different from the so-called 'smooth' reovirus, the ICP frame in all the turreted reoviruses (λ1 in MRV or VP3 in aquareovirus) is stabilized by a set of clamped subunits, such as σ2 in MRV or VP6 in aquareovirus. In fact, the 'clamp' protein has an additional role as a mediator bridging the inner core with the outer capsid, with the only exception in this sub-family being CVP. It has been shown that the 600 subunits of μ1 and σ3 (200 copies of the μ3σ3 heterohexamer) in orthoreovirus are arranged on an incomplete T=13 icosahedral lattice surrounding their ICPs. Further cryo-EM reconstructions at 9 Å resolution has revealed that the outer capsid of aquareovirus is composed of 200 trimers of VP5-VP7 heterodimers (Fig. 3A). Notably, the overall structure of aquareovirus VP5 appears to be very similar to that of μ1 in MRV when the crystal structure of μ1 and VP5 density map are fitted together; in particular, both the μ1N and μ1C domains fit well into the 9 Å cryo-EM Image (Cheng L P, et al., 2008). In fact, based on bioinformatics analyses, the N-terminal region at the Asn42-Pro43 amide bond (autolytic cleavage site) is highly conserved with that of μ1, suggesting that aquareovirus VP5 and MRV μ1 may share similar biological functions. However, there are few structural similarities (about 12% sequence identity) in the outermost protein s3 in MRV and VP7 in aquareovirus (Cheng L P, et al., 2008). This identity is also much lower than the 24% sequence identity between VP5 and μ1. Moreover, there is a distinctive difference in the outer capsid organization between MRV and aquareovirus, as MRV contains a hemagglutinin fiber protein s1 for cell attachment located at the distal end of the five-fold axis, but no such protein has been identified in aquareovirus (Fang Q, et al., 2005; Cheng L P et al., 2008, 2010). In the same way as the μ1 protein, VP5 has an interaction with turret VP1, clamp VP6, and outer shell VP7 protein. Given that s1 in MRV has been identified as interacting with a host cell acceptor, it has been suggested that aquareovirus may have different cellular acceptors from those of MRV. Beyond the interior framework, the structures of the dsRNA viruses also diverge, reflecting different mechanisms for viral maturation and entry.

Figure 3. Surface view and outer capsid protein structures of turreted and non-turreted rotaviruses. A: Surface view of intact aquareovirus (top left), the blue triangle outlines one VP5/VP7 heterohexamer (the analogue μ3s3 in MRV); the top right is VP5-VP7 complex in diagrammatic presentation, the three VP7 molecules(s3 in MRV) are located on the top, the three molecules of VP5 (μ1in MRV) are at the bottom. B: Cut-away view of the complete rotavirus virion is shown on the bottom left, and the cleaved VP4 (VP8* and VP5*) on the bottom right. The 3D images (A, B left) were reproduced from Cheng L P, et al., 2008. McClain B, et al., 2010.
As an example of smooth reovirus, rotavirus has a multi-shelled particle morphology (Estes M K, 2007). The outer protein layer of the rotavirus particle contains two protein components, VP4 and VP7. As shown in Fig. 3B, the two proteins (VP4 and VP7) form the outer-most layer of the rotavirus virion. VP7 has been identified as a glycoprotein, which is arranged as 260 trimers stabilized by Ca2+ ions, bound in the inter-subunit interfaces (Aoki S T, et al., 2009). VP7 trimers cap the trimeric pillars of VP6, and grip them with their amino-terminal arms. VP4 appears 60 trimeric spike-like projecting structures, which are required for infectivity. From a crystal structure analysis of VP7 (Aoki S T, et al., 2009) and high-resolution cryo-EM analysis of a VP7-recoated DLP (Chen J Z, et al., 2009), it is known that each VP7 trimer clamps onto an underlying VP6 trimer, through contacts made almost entirely by the VP7 N-terminal arms. The VP7 shell partly covers the base of the VP4 spike, and appears to lock VP4 onto the virion (Fig. 3B, right) (McClain B, et al., 2010). Trypsin cleavage of VP4 generates amino-terminal (VP8*) and carboxyl-terminal (VP5*) fragments of VP4, and primes virions for infectivity (Fig. 2B, right).
Based on the structural differences in the outer shell between turreted and smooth reoviruses, it is clear that outer capsid proteins are involved directly in virus and host cell interactions and exhibit a greater level of divergence.
-
To initiate infection of a target cell, non-enveloped viruses (those lacking a lipid-bilayer membrane) must disrupt the host cell membrane. To fulfill this purpose, non-enveloped dsRNA viruses have to transform from their environmentally stable state to a metastable state primed for membrane penetration to allow entry into cells. Based on the distinct multi-layered protein shell of reovirus, the specific protein that functions for virus penetration is generally maintained in the inactive state by protection from the outermost capsid 'chaperone' protein, such as σ3 in MRV or VP7 in aquareovirus (Liemmann S, et al., 2002; Cheng L P, et al., 2008). Upon initiation of infection, this viral penetration protein is usually converted from its dormant state to a metastable, primed state by proteolytic removal of its protector 'chaperone' protein.
Based on their particle organization, reoviruses seem to have an entry pathway different from that of enveloped viruses. Further, the different structural types of reovirusesmay have acquired distinct molecular mechanisms for carrying out this penetration step. Numerous studies have revealed that cell entry of reovirus is a multi-step process characterized by programmed disassembly of virions into at least two types of sub-virion particles, each with specialized roles in infection (Reinish K M, et al., 2000; Ivanovic T, et al., 2008). It has been reported previously that MRV attachment and penetration are functions of two distinct outer-layer proteins: σ1 and μ1, respectively (Dryden K A, et al., 1993). The fibers that project from the three copies of σ1 located at the icosahedral five-fold positions may perform attachment by interacting with host cells using a different pathway from that used by the nearby penetration protein μ1. The μ1 protein with an N-terminal myristoyl modification may carry out the penetration process during early inf ection. Except for the σ1 attachment protein pathway, the penetration protein of MRV μ1 needs to undergo a series of disassembly steps to activate its membrane-penetration machinery for delivery of its core into the cytoplasm. An autocatalytic cleavage separates the myristoylated N-terminal peptide, μ1N, from the rest of the μ1 protein (Liemann S, et al., 2002; Odegard A L, et al., 2004). The fragments known as δ and φ are the products of additional proteolysis events near the C-terminus. It has been suggested that μ1N and φ are released simultaneously from the particle, and act to form pores in the host cell membrane and recruit the reovirus core to these pores (Chandran K, et al., 2003; Ivanivic T, et al., 2008). The recent high-resolution infectious sub-viral particle (ISVP) reconstruction of aquareovirus at 3.3 Å resolution revealed that autoproteolysis of VP5 (the homolog of μ1) occurs at the Asn42-Pro43 bond during conversion from intact virion to ISVP (Zhang X, et al., 2010).
Sequence analysis and 3D reconstruction images suggest that the outer capsid protein VP5 in aquareovirus may have similar functions to those of μ1 in MRV, which has been suggested to be a penetration protein involved in cell entry (Cheng L P, et al., 2008). The 3.3 Å cryo-EM structure of aquareovirus ISVP revealed that the atomic structure of the primed VP5 has a 'Z' shape, and the VP5 subunits intertwine to form a trimer, which is consistent with the crystal structure reported previously for the orthoreovirus μ1 and σ3 hetero-hexamer (Liemann S, et al., 2002). The atomic model also indicates that conversion of VP5 from the dormant to the primed state is accompanied by autocleavage at the Asn42-Pro43 bond, probably facilitated by a nearby Lys84 that is normally held in check by a salt bridge with Glu76. In addition, the cryo-EM map of the ISVP directly reveals the density of a myristoyl group (a membrane insertion 'finger') sheltered in the hydrophobic pocket and covalently attached to the N-terminus. Release of the myristoyl group attached to the cleaved Gly2-Asn42 fragment corresponds to a second conversion from the primed ISVP to ISVP*, followed by interaction with the host membrane and consequent cell entry.
MRV infection is triggered by protease cleavage of the outer capsid protein μ13σ33 heterohexamers from the dormant particle. The resulting infectious subvirion particle is metastable. It can be prompted to undergo further disassembly to ISVP* in association with membrane penetration, and then led to activate its membrane-penetration machinery for delivery of its 'ICP payload' into the cytoplasm for further transcription and replication (Chandran K et al., 2002, 2003). It has been suggested that, during MRV infection, the endosomal proteases degrade σ3, leaving μ1 exposed. The next step is to cleave the μ1 protein, and generate μ1d and the C-terminal fragment φ, as diagrammed in Fig. 4A (Agosto M A, et al., 2008). In addition, the resulting ISVP further disassembles to form ISVP* by autocatalytic cleaving of μ1 to release the N-terminal myristoylated peptide μ1N and the larger fragment δ. ISVP* is still thought to be involved in association with membrane penetration. In MRV, conversion of ISVP* from ISVP and cleavage of μ1N from μ1 are both required for infectivity during cell entry (Odegard A L, et al., 2004).
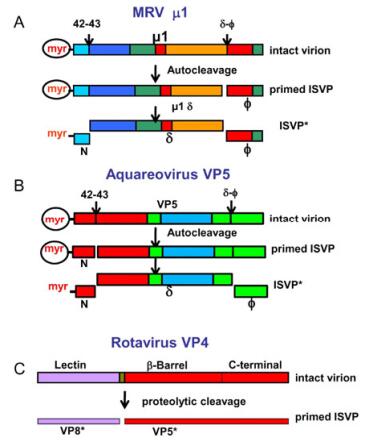
Figure 4. Conformational transformations of penetration proteins. A: Mammalian orthoreovirus (MRV) and B: Aquareovirus. The change of the penetration protein μ1/VP5 from a dormant state, to a metastable, primed infectious sub-viral particle (ISVP) state and subsequently to the ISVP* state. Colors indicate the domains in the μ1/VP5 subunits. C: Tryptic cleavage of VP4 generates VP8* (colored magenta in bar) and VP5* (red). Reproduced from Agosto M A, et al., 2008; Zhang X, et al., 2010a; Settembre E C, et al., 2011.
The current aquareovirus ISVP atomic model shows that VP5 is cleaved upon removal of VP7. This finding suggests that removal of the aquareovirus VP7 is accompanied by a conformational change and rearrangement of the penetration protein VP5 from the dormant to the primed ISVP state. The 3D atomic model of aquareovirus ISVP suggests that cleavage of VP5 is an autocleavage because the Asn42-Pro43 bond in the ISVP is located in the interior of the VP5 layer, which is inaccessible to external proteases (Zhang X, et al., 2010). Careful examination of the atomic structure for VP5 suggests a mechanism for Asn42-Pro43 autocleavage through nucleophilic attack. This hypothesis assumes that, during priming, this salt bridge will break to allow Lys84 to facilitate the autocleavage. Thus, the penetration protein VP5 can be cleaved into two fragments: the N-terminal myristoylated peptide VP5 and the remaining VP5 fragment. Following this step, the penetration protein may undergo an ordered disassembly that is triggered by several proteolysis events, ultimately leading to the release of hydrophobic fragments of the penetration protein (myristal-VP5N), and the remaining VP5 fragments, as shown schematically in Fig. 4B (Zhang X, et al., 2010a). The atomic structure reveals that autocleavage of the penetration protein with its N-terminal myristoylated peptide occurs during the conversion from the dormant to the primed state at an early st age of infection. This finding verified that the covalently linked myristoyl group tucks into a hydrophobic pocket at the base of each VP5 monomer, where it remains until it is released during the membrane penetration event (Liemann S, et al., 2002; Zhang X, et al., 2010a).
Removal of the outermost protein protector (σ3/VP7) results in an ISVP, which is thought to be primed for membrane penetration (Zhang X, et al., 2010a). Based on recent investigations into penetration proteins, either μ1 in MRV or VP5 in aquareovirus, the current model for reovirus membrane penetration suggests the following sequence of events in reovirus entry. First, there is removal of the 'chaperone' protein σ3/VP7 by chymotrypsin during endocytosis to cause a conformational change of μ1/VP5, which may promote autocleavage of μ1/VP5, and result in production of the metastable ISVP from the intact virion. The next steps may be involved in dynamic conformational changes and rearrangement of the penetration protein, coupled with proteolytic cleavage (Zhang L et al., 2006, 2009). Autocleavage is a critical step in cell entry for turreted reovirus, which changes the dormant virion to the primed ISVP. The late stage of membrane penetration is through conversion from the hydrophobic ISVP to a further disassembled particle ISVP, which induces the release of the myristoylated N-terminal peptide and the φ fragment.
It is clear that the particle organization of the outer capsid layer of smooth reovirus is rather different from that of spiked reovirus, but involvement of the proteolytic cleavage of VP4 protein in rotavirus early infection is similar to the mechanism in turreted reovirus. To gain high-level infectivity during cell entry, the spike VP4 protein of rotavirus has to be cleaved (Dormitzer P R, et al., 2004). During infectious entry, a sequence of molecular transformations from the triple-layered particle to its DLP occurs, resulting in delivery of the virus core particle into cells during the process of viral infection. VP8* and VP5* are two sub-fragments derived from proteolytic cleavage of VP4 in the initial infection step (Fig. 4C). The recent resolved full structure of rotavirus VP4 at near-atomic resolution extends the understanding of virus replication by combining previous structural work, and leads to a more specific model of the molecular events occurring during rotavirus penetration. The structure shows how the two s ub-fragments (VP8* and VP5*, cleaved from VP4) retain their association after proteolytic cleavage, revealing multiple structural roles for the β-barrel domain of VP5*, and specifying the interactions of VP4 with other capsid proteins (Settembre E C, et al., 2011). It also reveals that VP8* mediates cell attachment to terminal sialic acid moieties through the lectin domains at the tip of the spike. For entry of smooth virus, the lectin domains that interact with sialic acid moieties in the host cell are also a pathway to initiating infection. Orbivirus may have penetration machinery that is similar to rotavirus (Zhang X et al., 2010b).
-
Viruses of different structural classes have acquired distinct molecular mechanisms for carrying out infection and replication. The recent great progress in structural biology facilitated by high-resolution cryo-EM advances understanding of virus structures and related biological functions. The high-resolution reconstructions described in this review are substantial steps toward revealing the molecular mechanisms of non-enveloped virus assembly and entry.
Cell entry is a complex process involving one or more proteins. In general, for entry of a non-enveloped virus, the penetration proteins have to undergo a series of programmed cleavages associated with several conformational changes upon removing the outermost protein protector during the approach to the cell acceptor. Reconstructions of reoviruses at near-atomic resolution show that atomic models derived from cryo-EM images can provide valuable new information about virus assembly and entry. Even though only one conformational state of penetration proteins (e.g. μ1 in MRV, VP5 in aquareovirus or VP5* in rotavirus) has been disclosed by the current technical achievement, ongoing developments in both EM instrumentation and computation should extend the range of structures to image fields containing particles in more than one conformational state. High-resolution reconstructions of these structures will undoubtedly yield further insight into the entry mechanism of non-enveloped dsRNA viruses.
-
The work was supported by grants from the National Natural Science Foundation of China (31172434, 31372565).













 DownLoad:
DownLoad: